Epigenetic Alterations in Keratinocyte Carcinoma
- PMID: 33212152
- PMCID: PMC8068579
- DOI: 10.1016/j.jid.2020.10.018
Epigenetic Alterations in Keratinocyte Carcinoma
Abstract
Basal cell carcinoma (BCC) and squamous cell carcinoma (SCC) are both derived from epidermal keratinocytes but are phenotypically diverse. To improve the understanding of keratinocyte carcinogenesis, it is critical to understand epigenetic alterations, especially those that govern gene expression. We examined changes to the enhancer-associated histone acetylation mark H3K27ac by mapping matched tumor-normal pairs from 11 patients (five with BCC and six with SCC) undergoing Mohs surgery. Our analysis uncovered cancer-specific enhancers on the basis of differential H3K27ac peaks between matched tumor-normal pairs. We also uncovered biological pathways potentially altered in keratinocyte carcinoma, including enriched epidermal development and Wnt signaling pathways enriched in BCCs and enriched immune response and cell activation pathways in SCCs. We also observed enrichment of transcription factors that implicated SMAD and JDP2 in BCC pathogenesis and FOXP1 in SCC pathogenesis. On the basis of these findings, we prioritized three loci with putative regulation events (FGFR2 enhancer in BCC, intragenic regulation of FOXP1 in SCC, and WNT5A promoter in both subtypes) and validated our findings with published gene expression data. Our findings highlight unique and shared epigenetic alterations in histone modifications and potential regulators for BCCs and SCCs that likely impact the divergent oncogenic pathways, paving the way for targeted drug discoveries.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
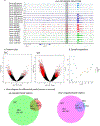



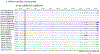


Comment in
-
From Enhancers to Keratinocyte Cancers.J Invest Dermatol. 2021 May;141(5):1134-1136. doi: 10.1016/j.jid.2020.11.008. J Invest Dermatol. 2021. PMID: 33888214 Free PMC article.
References
-
- Azzimonti B, Zavattaro E, Provasi M, Vidali M, Conca A, Catalano E, et al. Intense Foxp3+ CD25+ regulatory T-cell infiltration is associated with high-grade cutaneous squamous cell carcinoma and counterbalanced by CD8+/Foxp3+ CD25+ ratio. Br J Dermatol 2015;172(1):64–73. - PubMed
-
- Brown VL, Harwood CA, Crook T, Cronin JG, Kelsell DR, Proby CM. p16INK4a and p14ARF tumor suppressor genes are commonly inactivated in cutaneous squamous cell carcinoma. Journal of Investigative Dermatology 2004;122(5):1284–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

