Maternal obesity persistently alters cardiac progenitor gene expression and programs adult-onset heart disease susceptibility
- PMID: 33212270
- PMCID: PMC7720025
- DOI: 10.1016/j.molmet.2020.101116
Maternal obesity persistently alters cardiac progenitor gene expression and programs adult-onset heart disease susceptibility
Abstract
Objective: Heart disease risk can be programmed by intrauterine exposure to obesity. Dysregulating key transcription factors in cardiac progenitors can cause subsequent adult-onset heart disease. In this study, we investigated the transcriptional pathways that are altered in the embryonic heart and linked to heart disease risk in offspring exposed to obesity during pregnancy.
Methods: Female mice were fed an obesogenic diet and mated with males fed a control diet. Heart function and genome-wide gene expression were analyzed in adult offspring born to obese and lean mice at baseline and in response to stress. Cross-referencing with genes dysregulated genome-wide in cardiac progenitors from embryos of obese mice and human fetal hearts revealed the transcriptional events associated with adult-onset heart disease susceptibility.
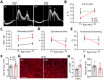
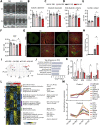
Results: We found that adult mice born to obese mothers develop mild heart dysfunction consistent with early stages of disease. Accordingly, hearts of these mice dysregulated genes controlling extracellular matrix remodeling, metabolism, and TGF-β signaling, known to control heart disease progression. These pathways were already dysregulated in cardiac progenitors in embryos of obese mice. Moreover, in response to cardiovascular stress, the heart of adults born to obese dams developed exacerbated myocardial remodeling and excessively activated regulators of cell-extracellular matrix interactions but failed to activate metabolic regulators. Expression of developmentally regulated genes was altered in cardiac progenitors of embryos of obese mice and human hearts of fetuses of obese donors. Accordingly, the levels of Nkx2-5, a key regulator of heart development, inversely correlated with maternal body weight in mice. Furthermore, Nkx2-5 target genes were dysregulated in cardiac progenitors and persistently in adult hearts born to obese mice and human hearts from pregnancies affected by obesity.
Conclusions: Obesity during pregnancy alters Nkx2-5-controlled transcription in differentiating cardiac progenitors and persistently in the adult heart, making the adult heart vulnerable to dysregulated stress responses.
Keywords: Cardiac progenitors; Fetal programming of heart disease; Gene expression; Heart disease susceptibility; Maternal obesity; Myocardial remodeling; Nkx2-5.
Copyright © 2020 The Authors. Published by Elsevier GmbH.. All rights reserved.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

