Transcriptomic Analysis of Staphylococcus xylosus in Solid Dairy Matrix Reveals an Aerobic Lifestyle Adapted to Rind
- PMID: 33212972
- PMCID: PMC7698506
- DOI: 10.3390/microorganisms8111807
Transcriptomic Analysis of Staphylococcus xylosus in Solid Dairy Matrix Reveals an Aerobic Lifestyle Adapted to Rind
Abstract
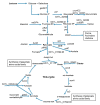
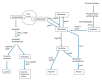
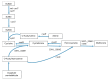
Staphylococcus xylosus is found in the microbiota of traditional cheeses, particularly in the rind of soft smeared cheeses. Despite its frequency, the molecular mechanisms allowing the growth and adaptation of S. xylosus in dairy products are still poorly understood. A transcriptomic approach was used to determine how the gene expression profile is modified during the fermentation step in a solid dairy matrix. S. xylosus developed an aerobic metabolism perfectly suited to the cheese rind. It overexpressed genes involved in the aerobic catabolism of two carbon sources in the dairy matrix, lactose and citrate. Interestingly, S. xylosus must cope with nutritional shortage such as amino acids, peptides, and nucleotides, consequently, an extensive up-regulation of genes involved in their biosynthesis was observed. As expected, the gene sigB was overexpressed in relation with general stress and entry into the stationary phase and several genes under its regulation, such as those involved in transport of anions, cations and in pigmentation were up-regulated. Up-regulation of genes encoding antioxidant enzymes and glycine betaine transport and synthesis systems showed that S. xylosus has to cope with oxidative and osmotic stresses. S. xylosus expressed an original system potentially involved in iron acquisition from lactoferrin.
Keywords: Staphylococcus; cheese model; nutrient shortage; osmotic stress; oxidative stress; physiology.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials

