A Novel Sub-Lineage of Chikungunya Virus East/Central/South African Genotype Indian Ocean Lineage Caused Sequential Outbreaks in Bangladesh and Thailand
- PMID: 33213040
- PMCID: PMC7698486
- DOI: 10.3390/v12111319
A Novel Sub-Lineage of Chikungunya Virus East/Central/South African Genotype Indian Ocean Lineage Caused Sequential Outbreaks in Bangladesh and Thailand
Abstract
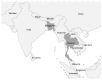
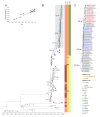
In recent decades, chikungunya virus (CHIKV) has become geographically widespread. In 2004, the CHIKV East/Central/South African (ECSA) genotype moved from Africa to Indian ocean islands and India followed by a large epidemic in Southeast Asia. In 2013, the CHIKV Asian genotype drove an outbreak in the Americas. Since 2016, CHIKV has re-emerged in the Indian subcontinent and Southeast Asia. In the present study, CHIKVs were obtained from Bangladesh in 2017 and Thailand in 2019, and their nearly full genomes were sequenced. Phylogenetic analysis revealed that the recent CHIKVs were of Indian Ocean Lineage (IOL) of genotype ECSA, similar to the previous outbreak. However, these CHIKVs were all clustered into a new distinct sub-lineage apart from the past IOL CHIKVs, and they lacked an alanine-to-valine substitution at position 226 of the E1 envelope glycoprotein, which enhances CHIKV replication in Aedes albopictus. Instead, all the re-emerged CHIKVs possessed mutations of lysine-to-glutamic acid at position 211 of E1 and valine-to-alanine at position 264 of E2. Molecular clock analysis suggested that the new sub-lineage CHIKV was introduced to Bangladesh around late 2015 and Thailand in early 2017. These results suggest that re-emerged CHIKVs have acquired different adaptations than the previous CHIKVs.
Keywords: Bangladesh; East/Central/South African genotype; Indian Ocean lineage; Thailand; chikungunya virus; molecular clock analysis; mosquito; outbreaks.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Figures

References
-
- Volk S.M., Chen R., Tsetsarkin K.A., Adams A.P., Garcia T.I., Sall A.A., Nasar F., Schuh A.J., Holmes E.C., Higgs S., et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J. Virol. 2010;84:6497–6504. doi: 10.1128/JVI.01603-09. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

