Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer
- PMID: 33213490
- PMCID: PMC7678301
- DOI: 10.1186/s13045-020-00991-2
Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer
Erratum in
-
Correction to: Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer.J Hematol Oncol. 2021 Feb 23;14(1):33. doi: 10.1186/s13045-021-01042-0. J Hematol Oncol. 2021. PMID: 33618743 Free PMC article. No abstract available.
Abstract
Background: Mounting evidence has demonstrated the vital importance of tumor-associated macrophages (TAMs) and exosomes in the formation of the premetastatic niche. However, the molecular mechanisms by which tumor-derived exosomal miRNAs interact with TAMs underlying premetastatic niche formation and colorectal cancer liver metastasis (CRLM) remain largely unknown.
Methods: Transmission electron microscopy and differential ultracentrifugation were used to verify the existence of exosomes. In vivo and in vitro assays were used to identify roles of exosomal miR-934. RNA pull-down assay, dual-luciferase reporter assay, etc. were applied to clarify the mechanism of exosomal miR-934 regulated the crosstalk between CRC cells and M2 macrophages.
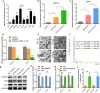
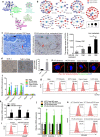
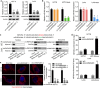
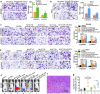
Results: In the present study, we first demonstrated the aberrant overexpression of miR-934 in colorectal cancer (CRC), especially in CRLM, and its correlation with the poor prognosis of CRC patients. Then, we verified that CRC cell-derived exosomal miR-934 induced M2 macrophage polarization by downregulating PTEN expression and activating the PI3K/AKT signaling pathway. Moreover, we revealed that hnRNPA2B1 mediated miR-934 packaging into exosomes of CRC cells and then transferred exosomal miR-934 into macrophages. Interestingly, polarized M2 macrophages could induce premetastatic niche formation and promote CRLM by secreting CXCL13, which activated a CXCL13/CXCR5/NFκB/p65/miR-934 positive feedback loop in CRC cells.
Conclusions: These findings indicate that tumor-derived exosomal miR-934 can promote CRLM by regulating the crosstalk between CRC cells and TAMs. These findings reveal a tumor and TAM interaction in the metastatic microenvironment mediated by tumor-derived exosomes that affects CRLM. The present study also provides a theoretical basis for secondary liver cancer.
Keywords: Colorectal cancer liver metastasis; Exosome; M2 macrophage polarization; Premetastatic niche; miR-934.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

