Genetic associations of in vivo pathology influence Alzheimer's disease susceptibility
- PMID: 33213512
- PMCID: PMC7678113
- DOI: 10.1186/s13195-020-00722-2
Genetic associations of in vivo pathology influence Alzheimer's disease susceptibility
Abstract
Introduction: Although the heritability of sporadic Alzheimer's disease (AD) is estimated to be 60-80%, addressing the genetic contribution to AD risk still remains elusive. More specifically, it remains unclear whether genetic variants are able to affect neurodegenerative brain features that can be addressed by in vivo imaging techniques.
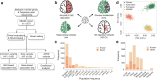
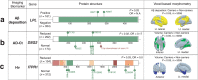
Methods: Targeted sequencing analysis of the coding and UTR regions of 132 AD susceptibility genes was performed. Neuroimaging data using 11C-Pittsburgh Compound B positron emission tomography (PET), 18F-fluorodeoxyglucose PET, and MRI that are available from the KBASE (Korean Brain Aging Study for Early Diagnosis and Prediction of Alzheimer's disease) cohort were acquired. A total of 557 participants consisted of 336 cognitively normal (CN) adults, 137 mild cognitive impairment (MCI), and 84 AD dementia (ADD) groups.
Results: We called 5391 high-quality single nucleotide variants (SNVs) on AD susceptibility genes and selected significant associations between variants and five in vivo AD pathologies: (1) amyloid β (Aβ) deposition, (2) AD-signature region cerebral glucose metabolism (AD-Cm), (3) posterior cingulate cortex (PCC) cerebral glucose metabolism (PCC-Cm), (4) AD-signature region cortical thickness (AD-Ct), and (5) hippocampal volume (Hv). The association analysis for common variants (allele frequency (AF) > 0.05) yielded several novel loci associated with Aβ deposition (PIWIL1-rs10848087), AD-Cm (NME8-rs2722372 and PSEN2-rs75733498), AD-Ct (PSEN1-rs7523) and, Hv (CASS4-rs3746625). Meanwhile, in a gene-based analysis for rare variants (AF < 0.05), cases carrying rare variants in LPL, FERMT2, NFAT5, DSG2, and ITPR1 displayed associations with the neuroimaging features. Exploratory voxel-based brain morphometry between the variant carriers and non-carriers was performed subsequently. Finally, we document a strong association of previously reported APOE variants with the in vivo AD pathologies and demonstrate that the variants exert a causal effect on AD susceptibility via neuroimaging features.
Conclusions: This study provides novel associations of genetic factors to Aβ accumulation and AD-related neurodegeneration to influence AD susceptibility.
Keywords: Alzheimer’s disease; Genetic association; In vivo AD pathologies; MRI; Neuroimaging; PET; Targeted panel sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–174. - PubMed
-
- Chouraki V, Seshadri S. Chapter five - Genetics of Alzheimer’s disease. In: Friedmann T, Dunlap JC, Goodwin SF, editors. Advances in Genetics. 87: Academic Press; 2014. p. 245–94. - PubMed
-
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278(16):1349–1356. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

