Network-based approach highlighting interplay among anti-hypertensives: target coding-genes: diseases
- PMID: 33214616
- PMCID: PMC7677320
- DOI: 10.1038/s41598-020-76605-1
Network-based approach highlighting interplay among anti-hypertensives: target coding-genes: diseases
Abstract
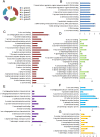
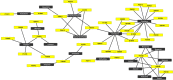
Elucidating the relation between the medicines: targets, targets: diseases and diseases: diseases are of fundamental significance as-is for societal benefit. Hypertension is one of the dangerous health conditions prevalent in society, is a risk factor for several other diseases if left untreated and anti-hypertensives (AHs) are the approved drugs to treat it. The goal of the study is to decipher the connection between hypertension with other health conditions, however, is challenging due to the large interactome. To fulfill the aim, the strategy involves prior clustering of the AHs into groups as per our previous method, followed by the analyzing functional association of the target coding-genes (tc-genes) and health conditions for each group. Following our recently published work where the AHs are clustered into six groups such that molecules having similar patterns come together, here, the distribution of molecular functions and the cellular components adopted by the tc-genes of each group are analyzed. The analyses indicate that kidney, heart, brain or lung related ailments are commonly associated with the tc-genes. The association of selective tc-genes to health conditions suggests a preference for certain health conditions despite many possibilities. Analyses of experimentally validated drug-drug combinations indicate the trend in successful AHs combinations. Clinically validated combinations bind different targets. Our study provides a promising methodology in a network-based approach that considers the influence of structural diversity of AHs to the functional perspective of tc-genes concerning the health conditions. The method could be extended to explore disease-disease relationships.
Conflict of interest statement
The author declares no competing interests.
Figures









Similar articles
-
Interrelation of cardiovascular risk factors with high albuminury among patients with arterial hypertension living in Mountain Shoriya.Ter Arkh. 2019 Mar 11;91(1):71-77. doi: 10.26442/00403660.2019.01.000033. Ter Arkh. 2019. PMID: 31090375
-
Data science-driven analyses of drugs inducing hypertension as an adverse effect.Mol Divers. 2021 May;25(2):801-810. doi: 10.1007/s11030-020-10059-5. Epub 2020 May 15. Mol Divers. 2021. PMID: 32415493
-
Pharmacogenetics of antihypertensive treatment.Vascul Pharmacol. 2006 Feb;44(2):107-18. doi: 10.1016/j.vph.2005.09.010. Epub 2005 Dec 13. Vascul Pharmacol. 2006. PMID: 16356784 Review.
-
Correlation of renin angiotensin system (RAS) candidate gene polymorphisms with response to Ramipril in patients with essential hypertension.J Postgrad Med. 2015 Jan-Mar;61(1):21-6. doi: 10.4103/0022-3859.147028. J Postgrad Med. 2015. PMID: 25511213 Free PMC article.
-
Unraveling the mechanisms of action of lactoferrin-derived antihypertensive peptides: ACE inhibition and beyond.Food Funct. 2015 Aug;6(8):2440-52. doi: 10.1039/c5fo00580a. Epub 2015 Jul 1. Food Funct. 2015. PMID: 26130265 Review.
References
-
- Kumar A, Tiwari A, Sharma A. Changing paradigm from one target one ligand towards multi-target directed ligand design for key drug targets of alzheimer disease: An important role of in silico methods in multi-target directed ligands design. Curr. Neuropharmacol. 2018;16(6):726–739. doi: 10.2174/1570159X16666180315141643. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

