Characterization of Plasmodium falciparum NEDD8 and identification of cullins as its substrates
- PMID: 33214620
- PMCID: PMC7677368
- DOI: 10.1038/s41598-020-77001-5
Characterization of Plasmodium falciparum NEDD8 and identification of cullins as its substrates
Abstract
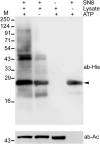
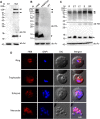
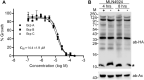
A variety of post-translational modifications of Plasmodium falciparum proteins, including phosphorylation and ubiquitination, are shown to have key regulatory roles during parasite development. NEDD8 is a ubiquitin-like modifier of cullin-RING E3 ubiquitin ligases, which regulates diverse cellular processes. Although neddylation is conserved in eukaryotes, it is yet to be characterized in Plasmodium and related apicomplexan parasites. We characterized P. falciparum NEDD8 (PfNEDD8) and identified cullins as its physiological substrates. PfNEDD8 is a 76 amino acid residue protein without the C-terminal tail, indicating that it can be readily conjugated. The wild type and mutant (Gly75Ala/Gly76Ala) PfNEDD8 were expressed in P. falciparum. Western blot of wild type PfNEDD8-expressing parasites indicated multiple high molecular weight conjugates, which were absent in the parasites expressing the mutant, indicating conjugation of NEDD8 through Gly76. Immunoprecipitation followed by mass spectrometry of wild type PfNEDD8-expressing parasites identified two putative cullins. Furthermore, we expressed PfNEDD8 in mutant S. cerevisiae strains that lacked endogenous NEDD8 (rub1Δ) or NEDD8 conjugating E2 enzyme (ubc12Δ). The PfNEDD8 immunoprecipitate also contained S. cerevisiae cullin cdc53, further substantiating cullins as physiological substrates of PfNEDD8. Our findings lay ground for investigation of specific roles and drug target potential of neddylation in malaria parasites.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
The Role of Neddylation in Malaria Parasites.DNA Cell Biol. 2024 Sep;43(9):426-429. doi: 10.1089/dna.2024.0120. Epub 2024 Jun 17. DNA Cell Biol. 2024. PMID: 38885136 Review.
-
The COP9 signalosome inhibits Cullin-RING E3 ubiquitin ligases independently of its deneddylase activity.Fly (Austin). 2018;12(2):118-126. doi: 10.1080/19336934.2018.1429858. Epub 2018 Feb 9. Fly (Austin). 2018. PMID: 29355077 Free PMC article.
-
Diverse and pivotal roles of neddylation in metabolism and immunity.FEBS J. 2021 Jul;288(13):3884-3912. doi: 10.1111/febs.15584. Epub 2020 Oct 20. FEBS J. 2021. PMID: 33025631 Review.
-
A lysine-to-arginine mutation on NEDD8 markedly reduces the activity of cullin RING E3 ligase through the impairment of neddylation cascades.Biochem Biophys Res Commun. 2015 Jun 12;461(4):653-8. doi: 10.1016/j.bbrc.2015.04.085. Epub 2015 Apr 24. Biochem Biophys Res Commun. 2015. PMID: 25918018
-
SENP8 limits aberrant neddylation of NEDD8 pathway components to promote cullin-RING ubiquitin ligase function.Elife. 2017 May 5;6:e24325. doi: 10.7554/eLife.24325. Elife. 2017. PMID: 28475037 Free PMC article.
Cited by
-
The emerging role of Deubiquitinases (DUBs) in parasites: A foresight review.Front Cell Infect Microbiol. 2022 Sep 27;12:985178. doi: 10.3389/fcimb.2022.985178. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36237424 Free PMC article. Review.
-
Functional characterisation of components in two Plasmodium falciparum Cullin-RING-Ligase complexes.Sci Rep. 2025 Jul 1;15(1):21359. doi: 10.1038/s41598-025-05342-0. Sci Rep. 2025. PMID: 40592953 Free PMC article.
-
The Skp1-Cullin1-FBXO1 complex is a pleiotropic regulator required for the formation of gametes and motile forms in Plasmodium berghei.Nat Commun. 2023 Mar 10;14(1):1312. doi: 10.1038/s41467-023-36999-8. Nat Commun. 2023. PMID: 36898988 Free PMC article.
-
Plasmodium falciparum contains functional SCF and CRL4 ubiquitin E3 ligases, and CRL4 is critical for cell division and membrane integrity.PLoS Pathog. 2024 Feb 28;20(2):e1012045. doi: 10.1371/journal.ppat.1012045. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38416790 Free PMC article.
-
Equine immunoglobulin fragment F(ab')2 displays high neutralizing capability against multiple SARS-CoV-2 variants.Clin Immunol. 2022 Apr;237:108981. doi: 10.1016/j.clim.2022.108981. Epub 2022 Mar 17. Clin Immunol. 2022. PMID: 35306171 Free PMC article.
References
-
- Walsh C. Posttranslational Modification of Proteins: Expanding Nature’s Inventory. New York: Roberts and Company Publishers; 2006.
-
- Organization WH. World Malaria Report 2019. Geneva: WHO; 2019.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

