Bacterial Operational Taxonomic Units Replace the Interactive Roles of Other Operational Taxonomic Units Under Strong Environmental Changes
- PMID: 33214767
- PMCID: PMC7604743
- DOI: 10.2174/1389202921999200716104355
Bacterial Operational Taxonomic Units Replace the Interactive Roles of Other Operational Taxonomic Units Under Strong Environmental Changes
Abstract
Background: Microorganisms are an important component of an aquatic ecosystem and play a critical role in the biogeochemical cycle which influences the circulation of the materials and maintains the balance in aquatic ecosystems.
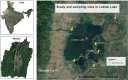
Objective: The seasonal variation along with the impact of anthropogenic activities, water quality, bacterial community composition and dynamics in the Loktak Lake, the largest freshwater lake of North East India, located in the Indo-Burma hotspot region was assessed during post-monsoon and winter season through metagenome analysis.
Methods: Five soil samples were collected during Post-monsoon and winter season from the Loktak Lake that had undergone different anthropogenic impacts. The metagenomic DNA of the soil samples was extracted using commercial metagenomic DNA extraction kits following the manufacturer's instruction. The extracted DNA was used to prepare the NGS library and sequenced in the Illumina MiSeq platform.
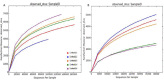
Results: Metagenomics analysis reveals Proteobacteria as the predominant community followed by Acidobacteria and Actinobacteria. The presence of these groups of bacteria indicates nitrogen fixation, oxidation of iron, sulfur, methane, and source of novel antibiotic candidates. The bacterial members belonging to different groups were involved in various biogeochemical processes, including fixation of carbon and nitrogen, producing streptomycin, gramicidin and perform oxidation of sulfur, sulfide, ammonia, and methane.
Conclusion: The outcome of this study provides a valuable dataset representing a seasonal profile across various land use and analysis, targeting at establishing an understanding of how the microbial communities vary across the land use and the role of keystone taxa. The findings may contribute to searches for microbial bio-indicators as biodiversity markers for improving the aquatic ecosystem of the Loktak Lake.
Keywords: Wetlands; bio-indicators; biogeochemical; keystone; land use; metagenomics; microbial diversity.
© 2020 Bentham Science Publishers.
Figures




References
-
- Graham E.B., Knelman J.E., Schindlbacher A., Siciliano S., Breulmann M., Yannarell A., Beman J.M., Abell G., Philippot L., Prosser J., Foulquier A., Yuste J.C., Glanville H.C., Jones D.L., Angel R., Salminen J., Newton R.J., Bürgmann H., Ingram L.J., Hamer U., Siljanen H.M.P., Peltoniemi K., Potthast K., Bañeras L., Hartmann M., Banerjee S., Yu R-Q., Nogaro G., Richter A., Koranda M., Castle S.C., Goberna M., Song B., Chatterjee A., Nunes O.C., Lopes A.R., Cao Y., Kaisermann A., Hallin S., Strickland M.S., Garcia-Pausas J., Barba J., Kang H., Isobe K., Papaspyrou S., Pastorelli R., Lagomarsino A., Lindström E.S., Basiliko N., Nemergut D.R. Microbes as engines of ecosystem function: when does community structure enhance predictions of ecosystem processes? Front. Microbiol. 2016;7:214. doi: 10.3389/fmicb.2016.00214. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
