Hypothetical Proteins as Predecessors of Long Non-coding RNAs
- PMID: 33214769
- PMCID: PMC7604745
- DOI: 10.2174/1389202921999200611155418
Hypothetical Proteins as Predecessors of Long Non-coding RNAs
Abstract
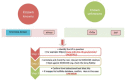
Hypothetical Proteins [HP] are the transcripts predicted to be expressed in an organism, but no evidence of it exists in gene banks. On the other hand, long non-coding RNAs [lncRNAs] are the transcripts that might be present in the 5' UTR or intergenic regions of the genes whose lengths are above 200 bases. With the known unknown [KU] regions in the genomes rapidly existing in gene banks, there is a need to understand the role of open reading frames in the context of annotation. In this commentary, we emphasize that HPs could indeed be the predecessors of lncRNAs.
Keywords: Hypothetical proteins; annotation; aptamers; functional genomics; lncRNA; transcripts.
© 2020 Bentham Science Publishers.
Figures

References
-
- Eisenstein E., Gilliland G.L., Herzberg O., Moult J., Orban J., Poljak R.J., Banerjei L., Richardson D., Howard A.J. Biological function made crystal clear-annotation of hypothetical proteins via structural genomics. Curr. Opin. Biotechnol. 2000;11(1):25–30. doi: 10.1016/S0958-1669(99)00063-4. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
