Comprehensive characterization of the ALMT and MATE families on Populus trichocarpa and gene co-expression network analysis of its members during aluminium toxicity and phosphate starvation stresses
- PMID: 33214973
- PMCID: PMC7658292
- DOI: 10.1007/s13205-020-02528-3
Comprehensive characterization of the ALMT and MATE families on Populus trichocarpa and gene co-expression network analysis of its members during aluminium toxicity and phosphate starvation stresses
Abstract
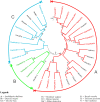
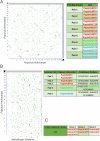
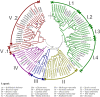
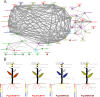
Aluminium (Al) toxicity and phosphate deficit on soils are some of the main problems of modern agriculture and are usually associated. Some plants are able to overcome these stresses through exuding organic acids on the rhizosphere, such as citrate and malate, which are exported by MATE (Multi drug and toxin extrusion) and ALMT (Aluminium-activated malate transporter) transporters, respectively. Despite its co-action on acidic soils, few studies explore these two families' correlation, especially on tree crops, therefore we performed a comprehensive description of MATE and ALMT families on Populus trichocarpa as a model species for arboreal plants. We found 20 and 56 putative members of ALMT and MATE families, respectively. Then, a gene co-expression network analysis was performed using broad transcriptomic data to analyze which members of each family were transcriptionally associated. Four independent networks were generated, one of which is composed of members putatively related to phosphate starvation and aluminum toxicity stresses. The PoptrALMT10 and PoptrMATE54 genes were selected from this network for a deeper analysis, which revealed that in roots under phosphate starvation stress the two genes have independent transcriptional profiles, however, on the aluminum toxicity stress they share some common correlations with other genes. The data presented here help on the description of these gene families, of which some members are potentially involved in plant responses to acid soil-related stresses and its exploration is an important step towards using this knowledge on breeding programs for P. trichocarpa and other tree crops.
Keywords: Acid soil; Aluminum toxicity; Gene regulation; Phosphorus starvation; Phylogenetic analyses; Synteny.
© King Abdulaziz City for Science and Technology 2020.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interest.
Figures



References
-
- Barcelo J, Poschenrieder C. Fast root growth responses, root exudates, and internal detoxification as clues to the mechanisms of aluminium toxicity and resistance: a review. Environ Exp Bot. 2002;48:75–92. doi: 10.1016/s0098-8472(02)00013-8. - DOI
LinkOut - more resources
Full Text Sources

