Using Machine Learning Modeling to Explore New Immune-Related Prognostic Markers in Non-Small Cell Lung Cancer
- PMID: 33215029
- PMCID: PMC7665579
- DOI: 10.3389/fonc.2020.550002
Using Machine Learning Modeling to Explore New Immune-Related Prognostic Markers in Non-Small Cell Lung Cancer
Abstract
Objective: To find new immune-related prognostic markers for non-small cell lung cancer (NSCLC).
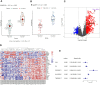
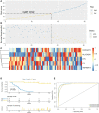
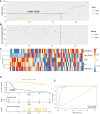
Methods: We found GSE14814 is related to NSCLC in GEO database. The non-small cell lung cancer observation (NSCLC-OBS) group was evaluated for immunity and divided into high and low groups for differential gene screening according to the score of immune evaluation. A single factor COX regression analysis was performed to select the genes related to prognosis. A prognostic model was constructed by machine learning, and test whether the model has a test efficacy for prognosis. A chip-in-chip non-small cell lung cancer chemotherapy (NSCLC-ACT) sample was used as a validation dataset for the same validation and prognostic analysis of the model. The coexpression genes of hub genes were obtained by pearson analysis and gene enrichment, function enrichment and protein interaction analysis. The tumor samples of patients with different clinical stages were detected by immunohistochemistry and the expression difference of prognostic genes in tumor tissues of patients with different stages was compared.
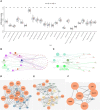
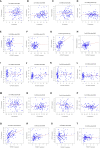
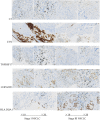
Results: By screening, we found that LYN, C3, COPG2IT1, HLA.DQA1, and TNFRSF17 is closely related to prognosis. After machine learning, we constructed the immune prognosis model from these 5 genes, and the model AUC values were greater than 0.9 at three time periods of 1, 3, and 5 years; the total survival period of the low-risk group was significantly better than that of the high-risk group. The results of prognosis analysis in ACT samples were consistent with OBS groups. The coexpression genes are mainly involved B cell receptor signaling pathway and are mainly enriched in apoptotic cell clearance. Prognostic key genes are highly correlated with PDCD1, PDCD1LG2, LAG3, and CTLA4 immune checkpoints. The immunohistochemical results showed that the expression of COPG2IT1 and HLA.DQA1 in stage III increased significantly and the expression of LYN, C3, and TNFRSF17 in stage III decreased significantly compared with that of stage I. The experimental results are consistent with the previous analysis.
Conclusion: LYN, C3, COPG2IT1, LA.DQA1, and NFRSF17 may be new immune markers to judge the prognosis of patients with non-small cell lung cancer.
Keywords: immune-related prognostic markers; immunohistochemistry; machine learning; model building; non-small cell lung cancer.
Copyright © 2020 Xu, Nie, He, Wang, Liao, Tu and Xiong.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

