Rare genetic variation at transcription factor binding sites modulates local DNA methylation profiles
- PMID: 33216750
- PMCID: PMC7679001
- DOI: 10.1371/journal.pgen.1009189
Rare genetic variation at transcription factor binding sites modulates local DNA methylation profiles
Abstract
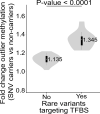
Although DNA methylation is the best characterized epigenetic mark, the mechanism by which it is targeted to specific regions in the genome remains unclear. Recent studies have revealed that local DNA methylation profiles might be dictated by cis-regulatory DNA sequences that mainly operate via DNA-binding factors. Consistent with this finding, we have recently shown that disruption of CTCF-binding sites by rare single nucleotide variants (SNVs) can underlie cis-linked DNA methylation changes in patients with congenital anomalies. These data raise the hypothesis that rare genetic variation at transcription factor binding sites (TFBSs) might contribute to local DNA methylation patterning. In this work, by combining blood genome-wide DNA methylation profiles, whole genome sequencing-derived SNVs from 247 unrelated individuals along with 133 predicted TFBS motifs derived from ENCODE ChIP-Seq data, we observed an association between the disruption of binding sites for multiple TFs by rare SNVs and extreme DNA methylation values at both local and, to a lesser extent, distant CpGs. While the majority of these changes affected only single CpGs, 24% were associated with multiple outlier CpGs within ±1kb of the disrupted TFBS. Interestingly, disruption of functionally constrained sites within TF motifs lead to larger DNA methylation changes at nearby CpG sites. Altogether, these findings suggest that rare SNVs at TFBS negatively influence TF-DNA binding, which can lead to an altered local DNA methylation profile. Furthermore, subsequent integration of DNA methylation and RNA-Seq profiles from cardiac tissues enabled us to observe an association between rare SNV-directed DNA methylation and outlier expression of nearby genes. In conclusion, our findings not only provide insights into the effect of rare genetic variation at TFBS on shaping local DNA methylation and its consequences on genome regulation, but also provide a rationale to incorporate DNA methylation data to interpret the functional role of rare variants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

