Coinfection by Severe Acute Respiratory Syndrome Coronavirus 2 and Influenza A(H1N1)pdm09 Virus Enhances the Severity of Pneumonia in Golden Syrian Hamsters
- PMID: 33216851
- PMCID: PMC7717201
- DOI: 10.1093/cid/ciaa1747
Coinfection by Severe Acute Respiratory Syndrome Coronavirus 2 and Influenza A(H1N1)pdm09 Virus Enhances the Severity of Pneumonia in Golden Syrian Hamsters
Abstract
Background: Clinical outcomes of the interaction between the co-circulating pandemic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and seasonal influenza viruses are unknown.
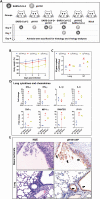
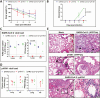
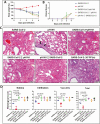
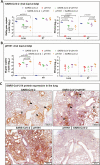
Methods: We established a golden Syrian hamster model coinfected by SARS-CoV-2 and mouse-adapted A(H1N1)pdm09 simultaneously or sequentially. The weight loss, clinical scores, histopathological changes, viral load and titer, and serum neutralizing antibody titer were compared with hamsters challenged by either virus.
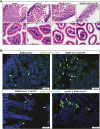
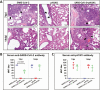
Results: Coinfected hamsters had more weight loss, more severe lung inflammatory damage, and tissue cytokine/chemokine expression. Lung viral load, infectious virus titers, and virus antigen expression suggested that hamsters were generally more susceptible to SARS-CoV-2 than to A(H1N1)pdm09. Sequential coinfection with A(H1N1)pdm09 one day prior to SARS-CoV-2 exposure resulted in a lower lung SARS-CoV-2 titer and viral load than with SARS-CoV-2 monoinfection, but a higher lung A(H1N1)pdm09 viral load. Coinfection also increased intestinal inflammation with more SARS-CoV-2 nucleoprotein expression in enterocytes. Simultaneous coinfection was associated with delay in resolution of lung damage, lower serum SARS-CoV-2 neutralizing antibody, and longer SARS-CoV-2 shedding in oral swabs compared to that of SARS-CoV-2 monoinfection.
Conclusions: Simultaneous or sequential coinfection by SARS-CoV-2 and A(H1N1)pdm09 caused more severe disease than monoinfection by either virus in hamsters. Prior A(H1N1)pdm09 infection lowered SARS-CoV-2 pulmonary viral loads but enhanced lung damage. Whole-population influenza vaccination for prevention of coinfection, and multiplex molecular diagnostics for both viruses to achieve early initiation of antiviral treatment for improvement of clinical outcome should be considered.
Keywords: COVID-19; SARS-CoV-2; coinfection; coronavirus; hamster; influenza; monoinfection.
© The Author(s) 2020. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures


Comment in
-
Coinfection, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), and Influenza: An Evolving Puzzle.Clin Infect Dis. 2021 Jun 15;72(12):e993-e994. doi: 10.1093/cid/ciaa1810. Clin Infect Dis. 2021. PMID: 33277660 Free PMC article. No abstract available.
Similar articles
-
Clinical and virological impact of single and dual infections with influenza A (H1N1) and SARS-CoV-2 in adult inpatients.PLoS Negl Trop Dis. 2021 Nov 29;15(11):e0009997. doi: 10.1371/journal.pntd.0009997. eCollection 2021 Nov. PLoS Negl Trop Dis. 2021. PMID: 34843492 Free PMC article.
-
Absence of Vaccine-enhanced Disease With Unexpected Positive Protection Against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by Inactivated Vaccine Given Within 3 Days of Virus Challenge in Syrian Hamster Model.Clin Infect Dis. 2021 Aug 2;73(3):e719-e734. doi: 10.1093/cid/ciab083. Clin Infect Dis. 2021. PMID: 33515458 Free PMC article.
-
Influenza A(H1N1)pdm09 Virus but Not Respiratory Syncytial Virus Interferes with SARS-CoV-2 Replication during Sequential Infections in Human Nasal Epithelial Cells.Viruses. 2022 Feb 15;14(2):395. doi: 10.3390/v14020395. Viruses. 2022. PMID: 35215988 Free PMC article.
-
Heterogeneity in transmissibility and shedding SARS-CoV-2 via droplets and aerosols.Elife. 2021 Apr 16;10:e65774. doi: 10.7554/eLife.65774. Elife. 2021. PMID: 33861198 Free PMC article.
-
Pathogenesis-directed therapy of 2019 novel coronavirus disease.J Med Virol. 2021 Mar;93(3):1320-1342. doi: 10.1002/jmv.26610. Epub 2020 Nov 10. J Med Virol. 2021. PMID: 33073355 Review.
Cited by
-
Effect of Structural Differences in Naringenin, Prenylated Naringenin, and Their Derivatives on the Anti-Influenza Virus Activity and Cellular Uptake of Their Flavanones.Pharmaceuticals (Basel). 2022 Nov 28;15(12):1480. doi: 10.3390/ph15121480. Pharmaceuticals (Basel). 2022. PMID: 36558931 Free PMC article.
-
An overview on viral interference during SARS-CoV-2 pandemic.Front Pediatr. 2023 Dec 21;11:1308105. doi: 10.3389/fped.2023.1308105. eCollection 2023. Front Pediatr. 2023. PMID: 38178911 Free PMC article. Review.
-
The Host Response to Influenza A Virus Interferes with SARS-CoV-2 Replication during Coinfection.J Virol. 2022 Aug 10;96(15):e0076522. doi: 10.1128/jvi.00765-22. Epub 2022 Jul 12. J Virol. 2022. PMID: 35862681 Free PMC article.
-
SARS-CoV-2 Interference of Influenza Virus Replication in Syrian Hamsters.J Infect Dis. 2022 Jan 18;225(2):282-286. doi: 10.1093/infdis/jiab587. J Infect Dis. 2022. PMID: 34875072 Free PMC article.
-
Outbreak of Influenza and Other Respiratory Viruses in Hospitalized Patients Alongside the SARS-CoV-2 Pandemic.Front Microbiol. 2022 Jun 13;13:902476. doi: 10.3389/fmicb.2022.902476. eCollection 2022. Front Microbiol. 2022. PMID: 35770154 Free PMC article.
References
-
- World Health Organization. Weekly epidemiological update—3 November 2020. Available at: https://www.who.int/docs/default-source/coronaviruse/situation-reports/w.... Accessed 11 June 2020.
-
- Li X, Sridhar S, Chan JF-W. The coronavirus disease 2019 pandemic: how does it spread and how do we stop it? Curr Opin HIV AIDS 2020; 15:328–35. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

