Membrane Tension Gates ERK-Mediated Regulation of Pluripotent Cell Fate
- PMID: 33217323
- PMCID: PMC7875115
- DOI: 10.1016/j.stem.2020.10.018
Membrane Tension Gates ERK-Mediated Regulation of Pluripotent Cell Fate
Abstract
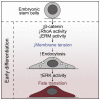
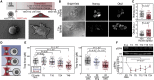
Cell fate transitions are frequently accompanied by changes in cell shape and mechanics. However, how cellular mechanics affects the instructive signaling pathways controlling cell fate is poorly understood. To probe the interplay between shape, mechanics, and fate, we use mouse embryonic stem cells (ESCs), which change shape as they undergo early differentiation. We find that shape change is regulated by a β-catenin-mediated decrease in RhoA activity and subsequent decrease in the plasma membrane tension. Strikingly, preventing a decrease in membrane tension results in early differentiation defects in ESCs and gastruloids. Decreased membrane tension facilitates the endocytosis of FGF signaling components, which activate ERK signaling and direct the exit from the ESC state. Increasing Rab5a-facilitated endocytosis rescues defective early differentiation. Thus, we show that a mechanically triggered increase in endocytosis regulates early differentiation. Our findings are of fundamental importance for understanding how cell mechanics regulates biochemical signaling and therefore cell fate.
Keywords: Beta-catenin; Cell fate choice; Cell surface mechanics; ERK; Embryonic stem cells; Endocytosis; Membrane tension; mechanical signalling; pluripotency.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





Comment in
-
Membrane Tension Locks In Pluripotency.Cell Stem Cell. 2021 Feb 4;28(2):175-176. doi: 10.1016/j.stem.2021.01.008. Cell Stem Cell. 2021. PMID: 33545073
References
-
- Avior Y., Sagi I., Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 2016;17:170–182. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

