Substrate Resistance to Traction Forces Controls Fibroblast Polarization
- PMID: 33217384
- PMCID: PMC7822749
- DOI: 10.1016/j.bpj.2020.10.043
Substrate Resistance to Traction Forces Controls Fibroblast Polarization
Abstract
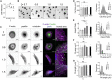
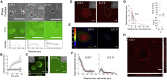
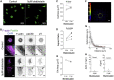
The mechanics of fibronectin-rich extracellular matrix regulate cell physiology in a number of diseases, prompting efforts to elucidate cell mechanosensing mechanisms at the molecular and cellular scale. Here, the use of fibronectin-functionalized silicone elastomers that exhibit considerable frequency dependence in viscoelastic properties unveiled the presence of two cellular processes that respond discreetly to substrate mechanical properties. Weakly cross-linked elastomers supported efficient focal adhesion maturation and fibroblast spreading because of an apparent stiff surface layer. However, they did not enable cytoskeletal and fibroblast polarization; elastomers with high cross-linking and low deformability were required for polarization. Our results suggest as an underlying reason for this behavior the inability of soft elastomer substrates to resist traction forces rather than a lack of sufficient traction force generation. Accordingly, mild inhibition of actomyosin contractility rescued fibroblast polarization even on the softer elastomers. Our findings demonstrate differential dependence of substrate physical properties on distinct mechanosensitive processes and provide a premise to reconcile previously proposed local and global models of cell mechanosensing.
Copyright © 2020 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Evans E.A., Calderwood D.A. Forces and bond dynamics in cell adhesion. Science. 2007;316:1148–1153. - PubMed
-
- Petridou N.I., Spiró Z., Heisenberg C.-P. Multiscale force sensing in development. Nat. Cell Biol. 2017;19:581–588. - PubMed
-
- Engler A.J., Sen S., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

