Substrate Specificity and Structural Modeling of Human Carboxypeptidase Z: A Unique Protease with a Frizzled-Like Domain
- PMID: 33217972
- PMCID: PMC7698808
- DOI: 10.3390/ijms21228687
Substrate Specificity and Structural Modeling of Human Carboxypeptidase Z: A Unique Protease with a Frizzled-Like Domain
Abstract
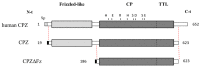
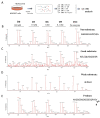
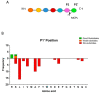
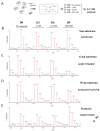
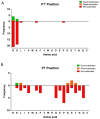
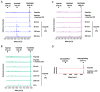
Metallocarboxypeptidase Z (CPZ) is a secreted enzyme that is distinguished from all other members of the M14 metallocarboxypeptidase family by the presence of an N-terminal cysteine-rich Frizzled-like (Fz) domain that binds Wnt proteins. Here, we present a comprehensive analysis of the enzymatic properties and substrate specificity of human CPZ. To investigate the enzymatic properties, we employed dansylated peptide substrates. For substrate specificity profiling, we generated two different large peptide libraries and employed isotopic labeling and quantitative mass spectrometry to study the substrate preference of this enzyme. Our findings revealed that CPZ has a strict requirement for substrates with C-terminal Arg or Lys at the P1' position. For the P1 position, CPZ was found to display specificity towards substrates with basic, small hydrophobic, or polar uncharged side chains. Deletion of the Fz domain did not affect CPZ activity as a carboxypeptidase. Finally, we modeled the structure of the Fz and catalytic domains of CPZ. Taken together, these studies provide the molecular elucidation of substrate recognition and specificity of the CPZ catalytic domain, as well as important insights into how the Fz domain binds Wnt proteins to modulate their functions.
Keywords: Wnt signaling; carboxypeptidase Z; cysteine rich domain; frizzled; growth factor; metallocarboxypeptidase; substrate specificity.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




Similar articles
-
Purification and characterization of human metallocarboxypeptidase Z.Biochem Biophys Res Commun. 1999 Mar 24;256(3):564-8. doi: 10.1006/bbrc.1999.0378. Biochem Biophys Res Commun. 1999. PMID: 10080937
-
Cloning, sequence analysis, and distribution of rat metallocarboxypeptidase Z.DNA Cell Biol. 1998 Apr;17(4):311-9. doi: 10.1089/dna.1998.17.311. DNA Cell Biol. 1998. PMID: 9570147
-
Carboxypeptidases from A to z: implications in embryonic development and Wnt binding.Cell Mol Life Sci. 2001 Nov;58(12-13):1790-804. doi: 10.1007/PL00000819. Cell Mol Life Sci. 2001. PMID: 11766880 Free PMC article. Review.
-
Substrate specificity of human metallocarboxypeptidase D: Comparison of the two active carboxypeptidase domains.PLoS One. 2017 Nov 13;12(11):e0187778. doi: 10.1371/journal.pone.0187778. eCollection 2017. PLoS One. 2017. PMID: 29131831 Free PMC article.
-
Disorders of FZ-CRD; insights towards FZ-CRD folding and therapeutic landscape.Mol Med. 2019 Dec 31;26(1):4. doi: 10.1186/s10020-019-0129-7. Mol Med. 2019. PMID: 31892318 Free PMC article. Review.
Cited by
-
Enhanced Production of ECM Proteins for Pharmaceutical Applications Using Mammalian Cells and Sodium Heparin Supplementation.Pharmaceutics. 2022 Oct 8;14(10):2138. doi: 10.3390/pharmaceutics14102138. Pharmaceutics. 2022. PMID: 36297573 Free PMC article.
-
By Increasing the Expression and Activation of STAT3, Sustained C5a Stimulation Increases the Proliferation, Migration, and Invasion of RCC Cells and Promotes the Growth of Transgrafted Tumors.Cancer Manag Res. 2021 Oct 4;13:7607-7621. doi: 10.2147/CMAR.S326352. eCollection 2021. Cancer Manag Res. 2021. PMID: 34675657 Free PMC article.
References
-
- Garcia-Pardo J., Grana-Montes R., Fernandez-Mendez M., Ruyra A., Roher N., Aviles F.X., Lorenzo J., Ventura S. Amyloid formation by human carboxypeptidase D transthyretin-like domain under physiological conditions. J. Biol. Chem. 2014;289:33783–33796. doi: 10.1074/jbc.M114.594804. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

