Discovery and Design of Novel Small Molecule GSK-3 Inhibitors Targeting the Substrate Binding Site
- PMID: 33218072
- PMCID: PMC7698860
- DOI: 10.3390/ijms21228709
Discovery and Design of Novel Small Molecule GSK-3 Inhibitors Targeting the Substrate Binding Site
Abstract
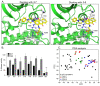
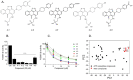
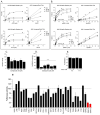
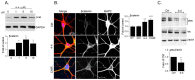
The serine/threonine kinase, GSK-3, is a promising drug discovery target for treating multiple pathological disorders. Most GSK-3 inhibitors that were developed function as ATP competitive inhibitors, with typical limitations in specificity, safety and drug-induced resistance. In contrast, substrate competitive inhibitors (SCIs), are considered highly selective, and more suitable for clinical practice. The development of SCIs has been largely neglected in the past because the ambiguous, undefined nature of the substrate-binding site makes them difficult to design. In this study, we used our previously described structural models of GSK-3 bound to SCI peptides, to design a pharmacophore model and to virtually screen the "drug-like" Zinc database (~6.3 million compounds). We identified leading hits that interact with critical binding elements in the GSK-3 substrate binding site and are chemically distinct from known GSK-3 inhibitors. Accordingly, novel GSK-3 SCI compounds were designed and synthesized with IC50 values of~1-4 μM. Biological activity of the SCI compound was confirmed in cells and in primary neurons that showed increased β-catenin levels and reduced tau phosphorylation in response to compound treatment. We have generated a new type of small molecule GSK-3 inhibitors and propose to use this strategy to further develop SCIs for other protein kinases.
Keywords: GSK-3; peptides; pharmacophore; small molecules; substrate competitive inhibitors; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

