Immune Phenotypes of Nasopharyngeal Cancer
- PMID: 33218184
- PMCID: PMC7699205
- DOI: 10.3390/cancers12113428
Immune Phenotypes of Nasopharyngeal Cancer
Abstract
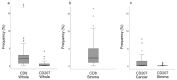
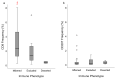
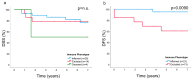
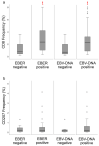
Nasopharyngeal cancer (NPC) features intralesional immune cells, but data are lacking on presence/distribution of T-cells and dendritic cells (DCs). Based on intralesional distribution of lymphocytes, a series of NPC biopsies (n = 48) were classified into "inflamed", "excluded", and "deserted" phenotypes. In addition, CD8+ T-cells and CD207+ DCs were quantified. The data were analyzed in relation to Epstein-Barr virus-encoded small RNA (EBER), Epstein-Barr virus (EBV) DNA, and survival. Separately, data on gene expression from a public database were analyzed. 61.7% of NPC lesions were "inflamed", 29.8% were "excluded", and 8.5% were "deserted". While CD8+ cells were present in cancer cell areas and in surrounding stroma, CD207+ cells were observed largely in cancer cell areas. High CD8+ T-cell presence was associated with EBV+ disease, but no such pattern was observed for CD207+ DCs. There was a difference in disease-free survival in favor of "inflamed" over "excluded" NPC. Gene expression analysis revealed differences between NPC and control tissue (e.g., with regard to interferon activity) as well as between subgroups of NPC based on CD8 expression (high vs. low). In conclusion, NPC lesions are heterogeneous with regard to distribution of CD8+ T-cells and CD207+ DCs. NPC can be classified into immune phenotypes that carry prognostic information. CD207+ DCs may represent a target for immunotherapy with potential to facilitate the antigen cross-presentation necessary to execute cytotoxic T-lymphocyte responses.
Keywords: CD207; CD8; cancer immune phenotype; quantification; survival.
Conflict of interest statement
The authors declare no conflict of interest. The funders have had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





References
-
- Wang Y.-Q., Zhang Y., Jiang W., Chen Y.-P., Xu S.-Y., Liu N., Zhao Y., Li L., Lei Y., Hong X.-H., et al. Development and validation of an immune checkpoint-based signature to predict prognosis in nasopharyngeal carcinoma using computational pathology analysis. J. Immunother. Cancer. 2019;7:1–11. doi: 10.1186/s40425-019-0752-4. - DOI - PMC - PubMed
-
- Chen Y.-P., Yin J.-H., Li W.-F., Li H.-J., Chen D.-P., Zhang C.-J., Lv J.-W., Wang Y.-Q., Li X.-M., Li J.-Y., et al. Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res. 2020:1–19. doi: 10.1038/s41422-020-0374-x. - DOI - PMC - PubMed
-
- Wang Y.-Q., Chen Y.-P., Zhang Y., Jiang W., Liu N., Yun J.-P., Sun Y., He Q.-M., Tang X.-R., Wen X., et al. Prognostic significance of tumor-infiltrating lymphocytes in nondisseminated nasopharyngeal carcinoma: A large-scale cohort study. Int. J. Cancer. 2018;142:2558–2566. doi: 10.1002/ijc.31279. - DOI - PubMed
-
- Fehres C.M., Duinkerken S., Bruijns S.C., Kalay H., Van Vliet S.J., Ambrosini M., De Gruijl T.D., Unger W.W., Garcia-Vallejo J.J., Van Kooyk Y. Langerin-mediated internalization of a modified peptide routes antigens to early endosomes and enhances cross-presentation by human Langerhans cells. Cell. Mol. Immunol. 2015;14:360–370. doi: 10.1038/cmi.2015.87. - DOI - PMC - PubMed
-
- Berg L.M.V.D., Cardinaud S., Van Der Aar A.M.G., Sprokholt J.K., De Jong M.A.W.P., Zijlstra-Willems E.M., Moris A., Geijtenbeek T.B.H. Langerhans Cell–Dendritic Cell Cross-Talk via Langerin and Hyaluronic Acid Mediates Antigen Transfer and Cross-Presentation of HIV-1. J. Immunol. 2015;195:1763–1773. doi: 10.4049/jimmunol.1402356. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

