Metagenomic Next-Generation Sequencing of Nasopharyngeal Specimens Collected from Confirmed and Suspect COVID-19 Patients
- PMID: 33219095
- PMCID: PMC7686804
- DOI: 10.1128/mBio.01969-20
Metagenomic Next-Generation Sequencing of Nasopharyngeal Specimens Collected from Confirmed and Suspect COVID-19 Patients
Abstract
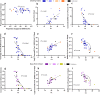
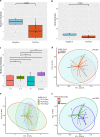
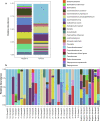
Metagenomic next-generation sequencing (mNGS) offers an agnostic approach for emerging pathogen detection directly from clinical specimens. In contrast to targeted methods, mNGS also provides valuable information on the composition of the microbiome and might uncover coinfections that may associate with disease progression and impact prognosis. To evaluate the use of mNGS for detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and/or other infecting pathogens, we applied direct Oxford Nanopore long-read third-generation metatranscriptomic and metagenomic sequencing. Nasopharyngeal (NP) swab specimens from 50 patients under investigation for CoV disease 2019 (COVID-19) were sequenced, and the data were analyzed by the CosmosID bioinformatics platform. Further, we characterized coinfections and the microbiome associated with a four-point severity index. SARS-CoV-2 was identified in 77.5% (31/40) of samples positive by RT-PCR, correlating with lower cycle threshold (Ct) values and fewer days from symptom onset. At the time of sampling, possible bacterial or viral coinfections were detected in 12.5% of SARS-CoV-2-positive specimens. A decrease in microbial diversity was observed among COVID-19-confirmed patients (Shannon diversity index, P = 0.0082; Chao richness estimate, P = 0.0097; Simpson diversity index, P = 0.018), and differences in microbial communities were linked to disease severity (P = 0.022). Furthermore, statistically significant shifts in the microbiome were identified among SARS-CoV-2-positive and -negative patients, in the latter of whom a higher abundance of Propionibacteriaceae (P = 0.028) and a reduction in the abundance of Corynebacterium accolens (P = 0.025) were observed. Our study corroborates the growing evidence that increased SARS-CoV-2 RNA detection from NP swabs is associated with the early stages rather than the severity of COVID-19. Further, we demonstrate that SARS-CoV-2 causes a significant change in the respiratory microbiome. This work illustrates the utility of mNGS for the detection of SARS-CoV-2, for diagnosing coinfections without viral target enrichment or amplification, and for the analysis of the respiratory microbiome.IMPORTANCE SARS-CoV-2 has presented a rapidly accelerating global public health crisis. The ability to detect and analyze viral RNA from minimally invasive patient specimens is critical to the public health response. Metagenomic next-generation sequencing (mNGS) offers an opportunity to detect SARS-CoV-2 from nasopharyngeal (NP) swabs. This approach also provides information on the composition of the respiratory microbiome and its relationship to coinfections or the presence of other organisms that may impact SARS-CoV-2 disease progression and prognosis. Here, using direct Oxford Nanopore long-read third-generation metatranscriptomic and metagenomic sequencing of NP swab specimens from 50 patients under investigation for COVID-19, we detected SARS-CoV-2 sequences by applying the CosmosID bioinformatics platform. Further, we characterized coinfections and detected a decrease in the diversity of the microbiomes in these patients. Statistically significant shifts in the microbiome were identified among COVID-19-positive and -negative patients, in the latter of whom a higher abundance of Propionibacteriaceae and a reduction in the abundance of Corynebacterium accolens were observed. Our study also corroborates the growing evidence that increased SARS-CoV-2 RNA detection from NP swabs is associated with the early stages of disease rather than with severity of disease. This work illustrates the utility of mNGS for the detection and analysis of SARS-CoV-2 from NP swabs without viral target enrichment or amplification and for the analysis of the respiratory microbiome.
Keywords: COVID-19; SARS-CoV-2; metagenomic next-generation sequencing; metagenomics; nasopharyngeal.
Copyright © 2020 Mostafa et al.
Figures
References
-
- Chen L, Liu W, Zhang Q, Xu K, Ye G, Wu W, Sun Z, Liu F, Wu K, Zhong B, Mei Y, Zhang W, Chen Y, Li Y, Shi M, Lan K, Liu Y. 2020. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect 9:313–319. doi: 10.1080/22221751.2020.1725399. - DOI - PMC - PubMed
-
- Moore SC, Penrice-Randal R, Alruwaili M, Dong X, Pullan ST, Carter D, Bewley K, Zhao Q, Sun Y, Hartley C, Zhou E, Solomon T, Beadsworth MBJ, Cruise J, Bogaert D, Crook DWT, Matthews DA, Davidson AD, Mahmood Z, Aljabr W, Druce J, Vipond RT, Ng L, Renia L, Openshaw P, Baillie JK, Carroll MW, Semple C, Turtle L, Hiscox JA. 2020. Amplicon based MinION sequencing of SARS-CoV-2 and metagenomic characterisation of nasopharyngeal swabs from patients with COVID-19. MedRxiv 2020.03.05.20032011.
-
- Manning JE, Bohl JA, Lay S, Chea S, Ly S, Sengdoeurn Y, Heng S, Vuthy C, Kalantar K, Ahyong V, Tan M, Sheu J, Tato CM, DeRisi J, Baril L, Dussart P, Duong V, Karlsson EA. 2020. Rapid metagenomic characterization of a case of imported COVID-19 in Cambodia. BioRxiv 2020.03.02.968818.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

