Gut microbiota-specific IgA+ B cells traffic to the CNS in active multiple sclerosis
- PMID: 33219152
- PMCID: PMC8043673
- DOI: 10.1126/sciimmunol.abc7191
Gut microbiota-specific IgA+ B cells traffic to the CNS in active multiple sclerosis
Abstract
Changes in gut microbiota composition and a diverse role of B cells have recently been implicated in multiple sclerosis (MS), a central nervous system (CNS) autoimmune disease. Immunoglobulin A (IgA) is a key regulator at the mucosal interface. However, whether gut microbiota shape IgA responses and what role IgA+ cells have in neuroinflammation are unknown. Here, we identify IgA-bound taxa in MS and show that IgA-producing cells specific for MS-associated taxa traffic to the inflamed CNS, resulting in a strong, compartmentalized IgA enrichment in active MS and other neuroinflammatory diseases. Unlike previously characterized polyreactive anti-commensal IgA responses, CNS IgA cross-reacts with surface structures on specific bacterial strains but not with brain tissue. These findings establish gut microbiota-specific IgA+ cells as a systemic mediator in MS and suggest a critical role of mucosal B cells during active neuroinflammation with broad implications for IgA as an informative biomarker and IgA-producing cells as an immune subset to harness for therapeutic interventions.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


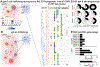
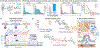
References
-
- Thompson AJ, Baranzini SE, Geurts J, Hemmer B, Ciccarelli O, Multiple sclerosis. Lancet 391, 1622–1636 (2018). - PubMed
-
- Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G, Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479, 538–541 (2011). - PubMed
-
- Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, Liu C, Klotz L, Stauffer U, Baranzini SE, Kümpfel T, Hohlfeld R, Krishnamoorthy G, Wekerle H, Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proceedings of the National Academy of Sciences of the United States of America 114, 10719–10724 (2017). - PMC - PubMed
-
- Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK, Hauser SL, Crabtree-Hartman E, Sand IK, Gacias M, Zhu Y, Casaccia P, Cree BAC, Knight R, Mazmanian SK, Baranzini SE, Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proceedings of the National Academy of Sciences of the United States of America 114, 10713–10718 (2017). - PMC - PubMed
-
- Cekanaviciute E, Pröbstel A-K, Thomann A, Runia TF, Casaccia P, Katz Sand I, Crabtree E, Singh S, Morrissey J, Barba P, Gomez R, Knight R, Mazmanian S, Graves J, Cree BAC, Zamvil SS, Baranzini SE, Multiple Sclerosis-Associated Changes in the Composition and Immune Functions of Spore-Forming Bacteria. mSystems 3, 353 (2018). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

