Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2
- PMID: 33219225
- PMCID: PMC7680145
- DOI: 10.1038/s41467-020-19097-x
Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2
Abstract
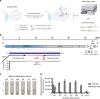
The COVID-19 pandemic has highlighted that new diagnostic technologies are essential for controlling disease transmission. Here, we develop SHINE (Streamlined Highlighting of Infections to Navigate Epidemics), a sensitive and specific diagnostic tool that can detect SARS-CoV-2 RNA from unextracted samples. We identify the optimal conditions to allow RPA-based amplification and Cas13-based detection to occur in a single step, simplifying assay preparation and reducing run-time. We improve HUDSON to rapidly inactivate viruses in nasopharyngeal swabs and saliva in 10 min. SHINE's results can be visualized with an in-tube fluorescent readout - reducing contamination risk as amplification reaction tubes remain sealed - and interpreted by a companion smartphone application. We validate SHINE on 50 nasopharyngeal patient samples, demonstrating 90% sensitivity and 100% specificity compared to RT-qPCR with a sample-to-answer time of 50 min. SHINE has the potential to be used outside of hospitals and clinical laboratories, greatly enhancing diagnostic capabilities.
Conflict of interest statement
C.A.F., P.C.S., and C.M. are inventors on patent PCT/US2018/022764, which covers the SHERLOCK and HUDSON technology for viral RNA detection held by the Broad Institute. J.A.-S., C.A.F., A.C.S., B.A.P., P.C.S., and C.M. are inventors on a pending patent application held by the Broad Institute (U.S. Provisional Patent Application No. 63/074,307). This pending application covers the SHINE technology and all designed sequences used in this work. J.E.L. consults for Sherlock Biosciences, Inc. P.C.S. is a co-founder of, shareholder in, and advisor to Sherlock Biosciences, Inc., as well as a Board member of and shareholder in Danaher Corporation. All other authors declare no competing interests.
Figures

Update of
-
Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2.bioRxiv [Preprint]. 2020 May 28:2020.05.28.119131. doi: 10.1101/2020.05.28.119131. bioRxiv. 2020. Update in: Nat Commun. 2020 Nov 20;11(1):5921. doi: 10.1038/s41467-020-19097-x. PMID: 32511415 Free PMC article. Updated. Preprint.
Similar articles
-
Integrated sample inactivation, amplification, and Cas13-based detection of SARS-CoV-2.bioRxiv [Preprint]. 2020 May 28:2020.05.28.119131. doi: 10.1101/2020.05.28.119131. bioRxiv. 2020. Update in: Nat Commun. 2020 Nov 20;11(1):5921. doi: 10.1038/s41467-020-19097-x. PMID: 32511415 Free PMC article. Updated. Preprint.
-
Performance of Abbott ID Now COVID-19 Rapid Nucleic Acid Amplification Test Using Nasopharyngeal Swabs Transported in Viral Transport Media and Dry Nasal Swabs in a New York City Academic Institution.J Clin Microbiol. 2020 Jul 23;58(8):e01136-20. doi: 10.1128/JCM.01136-20. Print 2020 Jul 23. J Clin Microbiol. 2020. PMID: 32471894 Free PMC article.
-
Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection.Biosens Bioelectron. 2020 Dec 1;169:112642. doi: 10.1016/j.bios.2020.112642. Epub 2020 Sep 20. Biosens Bioelectron. 2020. PMID: 32979593 Free PMC article.
-
Current and Perspective Diagnostic Techniques for COVID-19.ACS Infect Dis. 2020 Aug 14;6(8):1998-2016. doi: 10.1021/acsinfecdis.0c00365. Epub 2020 Aug 4. ACS Infect Dis. 2020. PMID: 32677821 Review.
-
Detection of COVID-19: A review of the current literature and future perspectives.Biosens Bioelectron. 2020 Oct 15;166:112455. doi: 10.1016/j.bios.2020.112455. Epub 2020 Jul 21. Biosens Bioelectron. 2020. PMID: 32739797 Free PMC article. Review.
Cited by
-
Next-generation CRISPR/Cas-based ultrasensitive diagnostic tools: current progress and prospects.RSC Adv. 2024 Oct 14;14(44):32411-32435. doi: 10.1039/d4ra04838e. eCollection 2024 Oct 9. RSC Adv. 2024. PMID: 39403159 Free PMC article. Review.
-
Research progress on nucleic acid detection and genome editing of CRISPR/Cas12 system.Mol Biol Rep. 2023 Apr;50(4):3723-3738. doi: 10.1007/s11033-023-08240-8. Epub 2023 Jan 17. Mol Biol Rep. 2023. PMID: 36648696 Free PMC article. Review.
-
CRISPR-Cas13a cascade-based viral RNA assay for detecting SARS-CoV-2 and its mutations in clinical samples.Sens Actuators B Chem. 2022 Jul 1;362:131765. doi: 10.1016/j.snb.2022.131765. Epub 2022 Mar 27. Sens Actuators B Chem. 2022. PMID: 35370361 Free PMC article.
-
CRISPR-based point-of-care diagnostics incorporating Cas9, Cas12, and Cas13 enzymes advanced for SARS-CoV-2 detection.J Biochem Mol Toxicol. 2022 Aug;36(8):e23113. doi: 10.1002/jbt.23113. Epub 2022 Jun 1. J Biochem Mol Toxicol. 2022. PMID: 35642647 Free PMC article. Review.
-
A chemical-enhanced system for CRISPR-Based nucleic acid detection.Biosens Bioelectron. 2021 Nov 15;192:113493. doi: 10.1016/j.bios.2021.113493. Epub 2021 Jul 9. Biosens Bioelectron. 2021. PMID: 34271398 Free PMC article.
References
-
- Kaplan, S. & Thomas, K. Despite promises, testing delays leave Americans ‘flying blind’. https://www.nytimes.com/2020/04/06/health/coronavirus-testing-us.html (2020).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

