Validation of a modified CDC assay and performance comparison with the NeuMoDx™ and DiaSorin® automated assays for rapid detection of SARS-CoV-2 in respiratory specimens
- PMID: 33220549
- PMCID: PMC7657030
- DOI: 10.1016/j.jcv.2020.104688
Validation of a modified CDC assay and performance comparison with the NeuMoDx™ and DiaSorin® automated assays for rapid detection of SARS-CoV-2 in respiratory specimens
Abstract
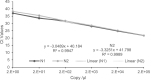
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19), has spread rapidly around the globe since it was first identified in December of 2019 in Wuhan, China. In a race to contain the infection, researchers and healthcare officials have developed several assays to help diagnose individuals with COVID-19. To help laboratories decide what assay to bring into testing lines, factors such as assay availability, cost, throughput, and TAT should be considered. Here we validated a modified version of the CDC assay and used it as a reference to evaluate the performance of the NeuMoDxTM SARS-CoV-2 and DiaSorin SimplexaTM Covid-19 Direct assays. In silico analysis and clinical sample testing showed that the primers/probes designed by the CDC were specific to the SARS-CoV-2 as they accurately detected all reactive samples with an assay LoD of 200 copies/mL. The performance of the three assays were analyzed using 159 nasopharyngeal swabs specimen tested within 1-5 days after routine testing. A 100 % agreement was observed between the commercial assays and the modified CDC SARS-CoV-2 assay. A deeper look at the Ct values showed no significant difference between NeuMoDx and the modified CDC SARS-CoV-2 assay, whereas DiaSorin had lower overall Ct values than the modified CDC SARS-CoV-2 assay. NeuMoDx and DiaSorin workflows were much easier to perform. NeuMoDx has the highest throughput and shortest TAT, whereas although the modified CDC SARS-CoV-2 assay has comparable throughput to DiaSorin, it has the longest hands-on time and highest TAT.
Keywords: COVID-19; Pandemic; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors report no declarations of interest.
Figures

Similar articles
-
Comparative study of four SARS-CoV-2 Nucleic Acid Amplification Test (NAAT) platforms demonstrates that ID NOW performance is impaired substantially by patient and specimen type.Diagn Microbiol Infect Dis. 2021 Jan;99(1):115200. doi: 10.1016/j.diagmicrobio.2020.115200. Epub 2020 Sep 3. Diagn Microbiol Infect Dis. 2021. PMID: 32980807 Free PMC article.
-
Comparison of Four Molecular In Vitro Diagnostic Assays for the Detection of SARS-CoV-2 in Nasopharyngeal Specimens.J Clin Microbiol. 2020 Jul 23;58(8):e00743-20. doi: 10.1128/JCM.00743-20. Print 2020 Jul 23. J Clin Microbiol. 2020. PMID: 32341143 Free PMC article.
-
Clinical performance and sample freeze-thaw stability of the cobas®6800 SARS-CoV-2 assay for the detection of SARS-CoV-2 in oro-/nasopharyngeal swabs and lower respiratory specimens.J Clin Virol. 2020 Dec;133:104686. doi: 10.1016/j.jcv.2020.104686. Epub 2020 Nov 8. J Clin Virol. 2020. PMID: 33221622 Free PMC article.
-
Detection of SARS-CoV-2 using real-time polymerase chain reaction in different clinical specimens: A critical review.Allergol Immunopathol (Madr). 2021 Jan 2;49(1):159-164. doi: 10.15586/aei.v49i1.60. eCollection 2021. Allergol Immunopathol (Madr). 2021. PMID: 33528945 Review.
-
Molecular Diagnostic Tools for the Detection of SARS-CoV-2.Int Rev Immunol. 2021;40(1-2):143-156. doi: 10.1080/08830185.2020.1871477. Epub 2021 Jan 13. Int Rev Immunol. 2021. PMID: 33439059 Review.
Cited by
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
-
Impact of rituximab on COVID-19 outcomes.Ann Hematol. 2021 Nov;100(11):2805-2812. doi: 10.1007/s00277-021-04662-1. Epub 2021 Sep 22. Ann Hematol. 2021. PMID: 34549309 Free PMC article.
-
Strategies for Scaling up SARS-CoV-2 Molecular Testing Capacity.Clin Lab Med. 2022 Jun;42(2):261-282. doi: 10.1016/j.cll.2022.02.006. Epub 2022 Mar 8. Clin Lab Med. 2022. PMID: 35636826 Free PMC article. Review.
-
An Overview of SARS-CoV-2 Molecular Diagnostics in Europe.Clin Lab Med. 2022 Jun;42(2):161-191. doi: 10.1016/j.cll.2022.02.005. Epub 2022 Mar 8. Clin Lab Med. 2022. PMID: 35636820 Free PMC article. Review.
-
Increasing diagnostic possibilities using the geneLEAD VIII platform for detection of SARS-CoV-2.J Virol Methods. 2021 Dec;298:114291. doi: 10.1016/j.jviromet.2021.114291. Epub 2021 Sep 23. J Virol Methods. 2021. PMID: 34562515 Free PMC article.
References
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. Epub 2020 Jan 24. - DOI - PMC - PubMed
-
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. Epub 2020 Jan 31. - DOI - PMC - PubMed
-
- Visseaux B., Le Hingrat Q., Collin G., Bouzid D., Lebourgeois S., Le Pluart D., Deconinck L., Lescure F.X., Lucet J.C., Bouadma L., Timsit J.F., Descamps D., Yazdanpanah Y., Casalino E., Houhou-Fidouh N. Evaluation of the QIAstat-Dx respiratory SARS-CoV-2 panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00630-20. - DOI - PMC - PubMed
-
- Nörz D., Fischer N., Schultze A., Kluge S., Mayer-Runge U., Aepfelbacher M., Pfefferle S., Lütgehetmann M. Clinical evaluation of a SARS-CoV-2 RT-PCR assay on a fully automated system for rapid on-demand testing in the hospital setting. J. Clin. Virol. 2020;128:104390. doi: 10.1016/j.jcv.2020.104390. Epub 2020 Apr 28. - DOI - PMC - PubMed
-
- Bordi L., Piralla A., Lalle E., Giardina F., Colavita F., Tallarita M., Sberna G., Novazzi F., Meschi S., Castilletti C., Brisci A., Minnucci G., Tettamanzi V., Baldanti F., Capobianchi M.R. Rapid and sensitive detection of SARS-CoV-2 RNA using the Simplexa™ COVID-19 direct assay. J. Clin. Virol. 2020;128:104416. doi: 10.1016/j.jcv.2020.104416. Epub 2020 May 4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

