Specific Guanosines in the HIV-2 Leader RNA are Essential for Efficient Viral Genome Packaging
- PMID: 33221337
- PMCID: PMC9168803
- DOI: 10.1016/j.jmb.2020.11.017
Specific Guanosines in the HIV-2 Leader RNA are Essential for Efficient Viral Genome Packaging
Abstract
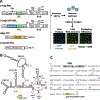
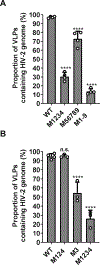
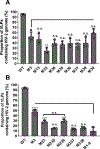
HIV-2, a human pathogen that causes acquired immunodeficiency syndrome, is distinct from the more prevalent HIV-1 in several features including its evolutionary history and certain aspects of viral replication. Like other retroviruses, HIV-2 packages two copies of full-length viral RNA during virus assembly and efficient genome encapsidation is mediated by the viral protein Gag. We sought to define cis-acting elements in the HIV-2 genome that are important for the encapsidation of full-length RNA into viral particles. Based on previous studies of murine leukemia virus and HIV-1, we hypothesized that unpaired guanosines in the 5' untranslated region (UTR) play an important role in Gag:RNA interactions leading to genome packaging. To test our hypothesis, we targeted 18 guanosines located in 9 sites within the HIV-2 5' UTR and performed substitution analyses. We found that mutating as few as three guanosines significantly reduce RNA packaging efficiency. However, not all guanosines examined have the same effect; instead, a hierarchical order exists wherein a primary site, a secondary site, and three tertiary sites are identified. Additionally, there are functional overlaps in these sites and mutations of more than one site can act synergistically to cause genome packaging defects. These studies demonstrate the importance of specific guanosines in HIV-2 5'UTR in mediating genome packaging. Our results also demonstrate an interchangeable and hierarchical nature of guanosine-containing sites, which was not previously established, thereby revealing key insights into the replication mechanisms of HIV-2.
Keywords: RNA; encapsidation; guanosine; retroviruses; untranslated region.
Published by Elsevier Ltd.
Figures

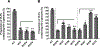
Similar articles
-
Elements in the 5' Untranslated Region of Viral RNA Important for HIV Gag Recognition and Cross-Packaging.Viruses. 2025 Apr 10;17(4):551. doi: 10.3390/v17040551. Viruses. 2025. PMID: 40284994 Free PMC article.
-
Unpaired Guanosines in the 5' Untranslated Region of HIV-1 RNA Act Synergistically To Mediate Genome Packaging.J Virol. 2020 Oct 14;94(21):e00439-20. doi: 10.1128/JVI.00439-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796062 Free PMC article.
-
Human immunodeficiency virus type 2 (HIV-2): packaging signal and associated negative regulatory element.Hum Gene Ther. 1995 Feb;6(2):177-84. doi: 10.1089/hum.1995.6.2-177. Hum Gene Ther. 1995. PMID: 7734518
-
HIV-1 RNA genome packaging: it's G-rated.mBio. 2024 Apr 10;15(4):e0086123. doi: 10.1128/mbio.00861-23. Epub 2024 Feb 27. mBio. 2024. PMID: 38411060 Free PMC article. Review.
-
Structural determinants and mechanism of HIV-1 genome packaging.J Mol Biol. 2011 Jul 22;410(4):609-33. doi: 10.1016/j.jmb.2011.04.029. J Mol Biol. 2011. PMID: 21762803 Free PMC article. Review.
Cited by
-
Human Retrovirus Genomic RNA Packaging.Viruses. 2022 May 19;14(5):1094. doi: 10.3390/v14051094. Viruses. 2022. PMID: 35632835 Free PMC article. Review.
-
MMTV RNA packaging requires an extended long-range interaction for productive Gag binding to packaging signals.PLoS Biol. 2024 Oct 3;22(10):e3002827. doi: 10.1371/journal.pbio.3002827. eCollection 2024 Oct. PLoS Biol. 2024. PMID: 39361708 Free PMC article.
-
Elements in the 5' Untranslated Region of Viral RNA Important for HIV Gag Recognition and Cross-Packaging.Viruses. 2025 Apr 10;17(4):551. doi: 10.3390/v17040551. Viruses. 2025. PMID: 40284994 Free PMC article.
-
Identification of a putative Gag binding site critical for feline immunodeficiency virus genomic RNA packaging.RNA. 2023 Dec 18;30(1):68-88. doi: 10.1261/rna.079840.123. RNA. 2023. PMID: 37914398 Free PMC article.
-
Single-Virion Analysis: A Method to Visualize HIV-1 Particle Content Using Fluorescence Microscopy.Methods Mol Biol. 2024;2807:77-91. doi: 10.1007/978-1-0716-3862-0_6. Methods Mol Biol. 2024. PMID: 38743222
References
-
- Peeters M, Jung M and Ayouba A (2013) The origin and molecular epidemiology of HIV. Expert Rev Anti Infect Ther, 11, 885–896. - PubMed
-
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM et al. (1999) Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature, 397, 436–441. - PubMed
-
- Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y et al. (2006) Human immunodeficiency viruses: SIV infection in wild gorillas. Nature, 444, 164. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

