Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen
- PMID: 33221508
- PMCID: PMC7661926
- DOI: 10.1016/j.bios.2020.112817
Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen
Abstract
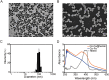
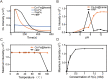
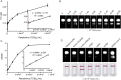
COVID-19 has evolved into a global pandemic. Early and rapid detection is crucial to control of the SARS-CoV-2 transmission. While representing the gold standard for early diagnosis, nucleic acid tests for SARS-CoV-2 are often complicated and time-consuming. Serological rapid antibody tests are characterized by high rates of false-negative diagnoses, especially during early infection. Here, we developed a novel nanozyme-based chemiluminescence paper assay for rapid and sensitive detection of SARS-CoV-2 spike antigen, which integrates nanozyme and enzymatic chemiluminescence immunoassay with the lateral flow strip. The core of our paper test is a robust Co-Fe@hemin-peroxidase nanozyme that catalyzes chemiluminescence comparable with natural peroxidase HRP and thus amplifies immune reaction signal. The detection limit for recombinant spike antigen of SARS-CoV-2 was 0.1 ng/mL, with a linear range of 0.2-100 ng/mL. Moreover, the sensitivity of test for pseudovirus could reach 360 TCID50/mL, which was comparable with ELISA method. The strip recognized SARS-CoV-2 antigen specifically, and there was no cross reaction with other coronaviruses or influenza A subtypes. This testing can be completed within 16 min, much shorter compared to the usual 1-2 h required for currently used nucleic acid tests. Furthermore, signal detection is feasible using the camera of a standard smartphone. Ingredients for nanozyme synthesis are simple and readily available, considerably lowering the overall cost. In conclusion, our paper test provides a high-sensitive point-of-care testing (POCT) approach for SARS-CoV-2 antigen detection, which should greatly facilitate early screening of SARS-CoV-2 infections, and considerably lower the financial burden on national healthcare resources.
Keywords: Antigen detection; Chemiluminescence; Nanozyme; Paper test; SARS-CoV-2.
Copyright © 2020. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K.J., Arunkumar G.A., Jurczyszak D., Polanco J., Bermudez-Gonzalez M., Kleiner G., Aydillo T., Miorin L., Fierer D.S., Lugo L.A., Kojic E.M., Stoever J., Liu S.T.H., Cunningham-Rundles C., Felgner P.L., Moran T., Garcia-Sastre A., Caplivski D., Cheng A.L.C., Kedzierska K., Vapalahti O., Hepojoki J.M., Simon V., Krammer F. Nat. Med. 2020 doi: 10.1038/s41591-020-0913-5. - DOI
-
- Deng H., Li X.L., Peng Q., Wang X., Chen J.P., Li Y.D. Angew. Chem. Int. Ed. 2005;44(18):2782–2785. - PubMed
-
- Deng J., Yang M., Wu J., Zhang W., Jiang X. Anal. Chem. 2018;90(15):9132–9137. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

