Fine-scale empirical data on niche divergence and homeolog expression patterns in an allopolyploid and its diploid progenitor species
- PMID: 33222195
- PMCID: PMC7986779
- DOI: 10.1111/nph.17101
Fine-scale empirical data on niche divergence and homeolog expression patterns in an allopolyploid and its diploid progenitor species
Erratum in
-
Corrigendum.New Phytol. 2021 Jul;231(1):500. doi: 10.1111/nph.17320. Epub 2021 Apr 25. New Phytol. 2021. PMID: 34060666 Free PMC article. No abstract available.
Abstract
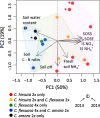
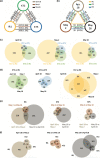
Polyploidization is pervasive in plants, but little is known about the niche divergence of wild allopolyploids (species that harbor polyploid genomes originating from different diploid species) relative to their diploid progenitor species and the gene expression patterns that may underlie such ecological divergence. We conducted a fine-scale empirical study on habitat and gene expression of an allopolyploid and its diploid progenitors. We quantified soil properties and light availability of habitats of an allotetraploid Cardamine flexuosa and its diploid progenitors Cardamine amara and Cardamine hirsuta in two seasons. We analyzed expression patterns of genes and homeologs (homeologous gene copies in allopolyploids) using RNA sequencing. We detected niche divergence between the allopolyploid and its diploid progenitors along water availability gradient at a fine scale: the diploids in opposite extremes and the allopolyploid in a broader range between diploids, with limited overlap with diploids at both ends. Most of the genes whose homeolog expression ratio changed among habitats in C. flexuosa varied spatially and temporally. These findings provide empirical evidence for niche divergence between an allopolyploid and its diploid progenitor species at a fine scale and suggest that divergent expression patterns of homeologs in an allopolyploid may underlie its persistence in diverse habitats.
Keywords: Cardamine; allopolyploid; homeolog expression; temporal fluctuation; transcriptome; water availability.
© 2020 The Authors New Phytologist © 2020 New Phytologist Foundation.
Figures




References
-
- Alexa A, Rahnenfuhrer J. 2016. topGO: enrichment analysis for gene ontology. R v.2.26.0. [WWW document] URL https://bioconductor.org/packages/topGO/.
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

