Notch 1 Is Involved in CD4+ T Cell Differentiation Into Th1 Subtype During Helicobacter pylori Infection
- PMID: 33224898
- PMCID: PMC7667190
- DOI: 10.3389/fcimb.2020.575271
Notch 1 Is Involved in CD4+ T Cell Differentiation Into Th1 Subtype During Helicobacter pylori Infection
Abstract
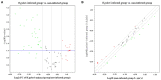
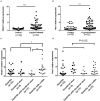
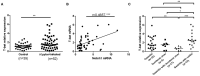
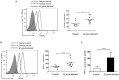
Helicobacter pylori infection induces CD4+ T differentiation cells into IFN-γ-producing Th1 cells. However, the details of mechanism underlying this process remain unclear. Notch signal pathway has been reported to regulate the differentiation of CD4+ T cells into Th1 subtype in many Th1-mediated inflammatory disorders but not yet in H. pylori infection. In the present study, the mRNA expression pattern of CD4+ T cells in H. pylori-infected patients differed from that of healthy control using Human Signal Transduction Pathway Finder RT2 Profiler PCR Array, and this alteration was associated with Notch signal pathway, as analyzed by Bioinformation. Quantitative real-time PCR showed that the mRNA expression of Notch1 and its target gene Hes-1 in CD4+ T cells of H. pylori-infected individuals increased compared with the healthy controls. In addition, the mRNA expression of Th1 master transcription factor T-bet and Th1 signature cytokine IFN-γ was both upregulated in H. pylori-infected individuals and positively correlated with Notch1 expression. The increased protein level of Notch1 and IFN-γ were also observed in H. pylori-infected individuals confirmed by flow cytometry and ELISA. In vitro, inhibition of Notch signaling decreased the mRNA expression of Notch1, Hes-1, T-bet, and IFN-γ, and reduced the protein levels of Notch1 and IFN-γ and the secretion of IFN-γ in CD4+ T cells stimulated by H. pylori. Collectively, this is the first evidence that Notch1 is upregulated and involved in the differentiation of Th1 cells during H. pylori infection, which will facilitate exploiting Notch1 as a therapeutic target for the control of H. pylori infection.
Keywords: CD4+ T cell; Helicobacter pylori; NOTCH1; Th1 cell; differentiation.
Copyright © 2020 Xie, Wen, Chen, Luo, Hu, Wu, Ye, Lin, Ning, Ning and Li.
Figures

Similar articles
-
Role of transcription factor T-bet expression by CD4+ cells in gastritis due to Helicobacter pylori in mice.Infect Immun. 2006 Aug;74(8):4673-84. doi: 10.1128/IAI.01887-05. Infect Immun. 2006. PMID: 16861655 Free PMC article.
-
The vast majority of gastric T cells are polarized to produce T helper 1 type cytokines upon antigenic stimulation despite the absence of Helicobacter pylori infection.J Gastroenterol. 1999 Oct;34(5):560-70. doi: 10.1007/s005350050373. J Gastroenterol. 1999. PMID: 10535482
-
Interleukin-12 drives the Th1 signaling pathway in Helicobacter pylori-infected human gastric mucosa.Infect Immun. 2007 Apr;75(4):1738-44. doi: 10.1128/IAI.01446-06. Epub 2007 Jan 12. Infect Immun. 2007. PMID: 17220306 Free PMC article.
-
The role of T helper 1-cell response in Helicobacter pylori-infection.Microb Pathog. 2018 Oct;123:1-8. doi: 10.1016/j.micpath.2018.06.033. Epub 2018 Jun 21. Microb Pathog. 2018. PMID: 29936093 Review.
-
[Immune response to an H. pylori infection in the stomach].Nihon Rinsho. 1999 Jan;57(1):23-31. Nihon Rinsho. 1999. PMID: 10036932 Review. Japanese.
Cited by
-
The potential value of Notch1 and DLL1 in the diagnosis and prognosis of patients with active TB.Front Immunol. 2023 Mar 28;14:1134123. doi: 10.3389/fimmu.2023.1134123. eCollection 2023. Front Immunol. 2023. PMID: 37063841 Free PMC article.
-
Helicobacter pylori promotes gastric cancer progression through the tumor microenvironment.Appl Microbiol Biotechnol. 2022 Jun;106(12):4375-4385. doi: 10.1007/s00253-022-12011-z. Epub 2022 Jun 20. Appl Microbiol Biotechnol. 2022. PMID: 35723694 Review.
-
Notch Signaling Ligand Jagged1 Enhances Macrophage-Mediated Response to Helicobacter pylori.Front Microbiol. 2021 Jul 8;12:692832. doi: 10.3389/fmicb.2021.692832. eCollection 2021. Front Microbiol. 2021. PMID: 34305857 Free PMC article.
-
Immune Biology and Persistence of Helicobacter pylori in Gastric Diseases.Curr Top Microbiol Immunol. 2023;444:83-115. doi: 10.1007/978-3-031-47331-9_4. Curr Top Microbiol Immunol. 2023. PMID: 38231216
-
The Recombinant Eg.P29-Mediated miR-126a-5p Promotes the Differentiation of Mouse Naive CD4+ T Cells via DLK1-Mediated Notch1 Signal Pathway.Front Immunol. 2022 Feb 8;13:773276. doi: 10.3389/fimmu.2022.773276. eCollection 2022. Front Immunol. 2022. PMID: 35211114 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

