Measures of Neural Similarity
- PMID: 33225218
- PMCID: PMC7671987
- DOI: 10.1007/s42113-019-00068-5
Measures of Neural Similarity
Abstract
One fundamental question is what makes two brain states similar. For example, what makes the activity in visual cortex elicited from viewing a robin similar to a sparrow? One common assumption in fMRI analysis is that neural similarity is described by Pearson correlation. However, there are a host of other possibilities, including Minkowski and Mahalanobis measures, with each differing in its mathematical, theoretical, and neural computational assumptions. Moreover, the operable measures may vary across brain regions and tasks. Here, we evaluated which of several competing similarity measures best captured neural similarity. Our technique uses a decoding approach to assess the information present in a brain region, and the similarity measures that best correspond to the classifier's confusion matrix are preferred. Across two published fMRI datasets, we found the preferred neural similarity measures were common across brain regions but differed across tasks. Moreover, Pearson correlation was consistently surpassed by alternatives.
Keywords: Machine learning; Neural coding; Neural similarity; fMRI.
© The Author(s) 2019.
Conflict of interest statement
Conflict of InterestsThe authors declare no competing financial interests.
Figures




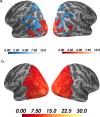
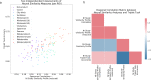
References
-
- Allefeld C, Haynes JD. Searchlight-based multi-voxel pattern analysis of fMRI by cross-validated MANOVA. NeuroImage. 2014;89:345–357. - PubMed
-
- Bartlett MS. The effect of standardization on a χ2 approximation in factor analysis. Biometrika. 1951;38(3/4):337–344.
-
- Bates D, Maechler M, Bolker B, Walker S. lme4: linear mixed-effects models using Eigen and S4. R package version. 2014;1(7):1–23.
Grants and funding
LinkOut - more resources
Full Text Sources
