Cardiac Pressure Overload Decreases ETV1 Expression in the Left Atrium, Contributing to Atrial Electrical and Structural Remodeling
- PMID: 33225722
- PMCID: PMC8449308
- DOI: 10.1161/CIRCULATIONAHA.120.048121
Cardiac Pressure Overload Decreases ETV1 Expression in the Left Atrium, Contributing to Atrial Electrical and Structural Remodeling
Erratum in
-
Correction to: Cardiac Pressure Overload Decreases ETV1 Expression in the Left Atrium, Contributing to Atrial Electrical and Structural Remodeling.Circulation. 2021 Feb 23;143(8):e745. doi: 10.1161/CIR.0000000000000951. Epub 2021 Feb 22. Circulation. 2021. PMID: 33617316 No abstract available.
Abstract
Background: Elevated intracardiac pressure attributable to heart failure induces electrical and structural remodeling in the left atrium (LA) that begets atrial myopathy and arrhythmias. The underlying molecular pathways that drive atrial remodeling during cardiac pressure overload are poorly defined. The purpose of this study is to characterize the response of the ETV1 (ETS translocation variant 1) signaling axis in the LA during cardiac pressure overload in humans and mouse models and explore the role of ETV1 in atrial electrical and structural remodeling.
Methods: We performed gene expression profiling in 265 left atrial samples from patients who underwent cardiac surgery. Comparative gene expression profiling was performed between 2 murine models of cardiac pressure overload, transverse aortic constriction banding and angiotensin II infusion, and a genetic model of Etv1 cardiomyocyte-selective knockout (Etv1f/fMlc2aCre/+).
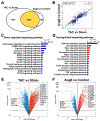
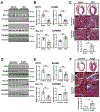
Results: Using the Cleveland Clinic biobank of human LA specimens, we found that ETV1 expression is decreased in patients with reduced ejection fraction. Consistent with its role as an important mediator of the NRG1 (Neuregulin 1) signaling pathway and activator of rapid conduction gene programming, we identified a direct correlation between ETV1 expression level and NRG1, ERBB4, SCN5A, and GJA5 levels in human LA samples. In a similar fashion to patients with heart failure, we showed that left atrial ETV1 expression is downregulated at the RNA and protein levels in murine pressure overload models. Comparative analysis of LA RNA sequencing datasets from transverse aortic constriction and angiotensin II-treated mice showed a high Pearson correlation, reflecting a highly ordered process by which the LA undergoes electrical and structural remodeling. Cardiac pressure overload produced a consistent downregulation of ErbB4, Etv1, Scn5a, and Gja5 and upregulation of profibrotic gene programming, which includes Tgfbr1/2, Igf1, and numerous collagen genes. Etv1f/fMlc2aCre/+ mice displayed atrial conduction disease and arrhythmias. Correspondingly, the LA from Etv1f/fMlc2aCre/+ mice showed downregulation of rapid conduction genes and upregulation of profibrotic gene programming, whereas analysis of a gain-of-function ETV1 RNA sequencing dataset from neonatal rat ventricular myocytes transduced with Etv1 showed reciprocal changes.
Conclusions: ETV1 is downregulated in the LA during cardiac pressure overload, contributing to both electrical and structural remodeling.
Keywords: atrial fibrillation; atrial remodeling; heart failure; signal transduction; transcription factors.
Figures





References
-
- Jessup M, Abraham WT, Casey DE, Feldman AM, Francis GS, Ganiats TG, Konstam MA, Mancini DM, Rahko PS, Silver MA, et al.2009 focused update: ACCF/AHA Guidelines for the Diagnosis and Management of Heart Failure in Adults: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines: developed in collaboration with the International Society for Heart and Lung Transplantation. Circulation. 2009;119:1977–2016. doi:10.1161/CIRCULATIONAHA.109.192064 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

