The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation
- PMID: 33225938
- PMCID: PMC7682012
- DOI: 10.1186/s12943-020-01272-9
The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation
Abstract
Background: Circular RNA (circRNAs) and hypoxia have been found to play the key roles in the pathogenesis and progression of cancer including colorectal cancer (CRC). However, the expressions and functions of the specific circRNAs in regulating hypoxia-involved CRC metastasis, and the circRNAs that are relevant to regulate HIF-1α levels in CRC remain elusive.
Methods: qRT-PCR was used to detect the expression of circRNAs and mRNA in CRC cells and tissues. Fluorescence in situ hybridization (FISH) was used to analyze the location of circ-ERBIN. Function-based experiments were performed using circ-ERBIN overexpression and knockdown cell lines in vitro and in vivo, including CCK8, colony formation, EdU assay, transwell, tumor growth and metastasis models. Mechanistically, luciferase reporter assay, western blots and immunohistochemical stainings were performed.
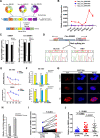
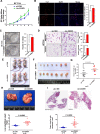
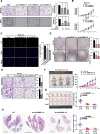
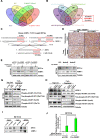
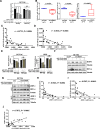
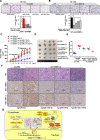
Results: Circ-Erbin was highly expressed in the CRC cells and Circ-Erbin overexpression facilitated the proliferation, migration and metastasis of CRC in vitro and in vivo. Notably, circ-Erbin overexpression significantly promoted angiogenesis by increasing the expression of hypoxia induced factor (HIF-1α) in CRC. Mechanistically, circ-Erbin accelerated a cap-independent protein translation of HIF-1α in CRC cells as the sponges of miR-125a-5p and miR-138-5p, which synergistically targeted eukaryotic translation initiation factor 4E binding protein 1(4EBP-1).
Conclusions: Our findings uncover a key mechanism for circ-Erbin mediated HIF-1α activation by miR-125a-5p-5p/miR-138-5p/4EBP-1 axis and circ-ERBIN is a potential target for CRC treatment.
Keywords: Circular RNA; Colorectal cancer; hypoxia; miRNA sponge.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures

Similar articles
-
Circular RNA circ_0007142 regulates cell proliferation, apoptosis, migration and invasion via miR-455-5p/SGK1 axis in colorectal cancer.Anticancer Drugs. 2021 Jan 1;32(1):22-33. doi: 10.1097/CAD.0000000000000992. Anticancer Drugs. 2021. PMID: 32889894
-
Circ_0000395 promotes cell growth, metastasis and oxaliplatin resistance by regulating miR-153-5p/MYO6 in colorectal cancer.Pathol Res Pract. 2024 Aug;260:155476. doi: 10.1016/j.prp.2024.155476. Epub 2024 Jul 17. Pathol Res Pract. 2024. PMID: 39038387
-
Knockdown of circular RNA circ-FARSA restricts colorectal cancer cell growth through regulation of miR-330-5p/LASP1 axis.Arch Biochem Biophys. 2020 Aug 15;689:108434. doi: 10.1016/j.abb.2020.108434. Epub 2020 May 29. Arch Biochem Biophys. 2020. PMID: 32473899
-
Interactions Between Non-Coding RNAs and HIF-1alpha in the Context of Colorectal Cancer.Biomolecules. 2025 Apr 1;15(4):510. doi: 10.3390/biom15040510. Biomolecules. 2025. PMID: 40305214 Free PMC article. Review.
-
Circ_CDR1as: A circular RNA with roles in the carcinogenesis.Pathol Res Pract. 2022 Aug;236:153968. doi: 10.1016/j.prp.2022.153968. Epub 2022 Jun 1. Pathol Res Pract. 2022. PMID: 35667198 Review.
Cited by
-
MiR-138-5p suppresses the progression of lung cancer by targeting SNIP1.Thorac Cancer. 2023 Feb;14(6):612-623. doi: 10.1111/1759-7714.14791. Epub 2023 Jan 3. Thorac Cancer. 2023. PMID: 36597175 Free PMC article.
-
Hypoxia-responsive circRNAs: A novel but important participant in non-coding RNAs ushered toward tumor hypoxia.Cell Death Dis. 2022 Aug 1;13(8):666. doi: 10.1038/s41419-022-05114-y. Cell Death Dis. 2022. PMID: 35915091 Free PMC article. Review.
-
Non-coding RNAs as Biomarkers for Survival in Colorectal Cancer Patients.Curr Aging Sci. 2024;17(1):5-15. doi: 10.2174/1874609816666230202101054. Curr Aging Sci. 2024. PMID: 36733201 Review.
-
Mechanism underlying circRNA dysregulation in the TME of digestive system cancer.Front Immunol. 2022 Sep 27;13:951561. doi: 10.3389/fimmu.2022.951561. eCollection 2022. Front Immunol. 2022. PMID: 36238299 Free PMC article. Review.
-
Deregulation of circRNA hsa_circ_0009109 promotes tumor growth and initiates autophagy by sponging miR-544a-3p in gastric cancer.Gastroenterol Rep (Oxf). 2024 Feb 28;12:goae008. doi: 10.1093/gastro/goae008. eCollection 2024. Gastroenterol Rep (Oxf). 2024. PMID: 38425655 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

