Histone modifications associated with gene expression and genome accessibility are dynamically enriched at Plasmodium falciparum regulatory sequences
- PMID: 33225957
- PMCID: PMC7682024
- DOI: 10.1186/s13072-020-00365-5
Histone modifications associated with gene expression and genome accessibility are dynamically enriched at Plasmodium falciparum regulatory sequences
Abstract
Background: The malaria parasite Plasmodium falciparum has an unusually euchromatic genome with poorly conserved positioning of nucleosomes in intergenic sequences and poorly understood mechanisms of gene regulation. Variant histones and histone modifications determine nucleosome stability and recruit trans factors, but their combinatorial contribution to gene regulation is unclear.
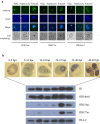
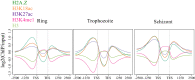
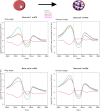
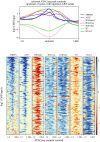
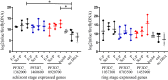
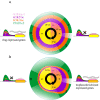
Results: Here, we show that the histone H3 acetylations H3K18ac and H3K27ac and the variant histone Pf H2A.Z are enriched together at regulatory sites upstream of genes. H3K18ac and H3K27ac together dynamically mark regulatory regions of genes expressed during the asexual life cycle. In contrast, H3K4me1 is depleted in intergenic sequence and dynamically depleted upstream of expressed genes. The temporal pattern of H3K27ac and H3K18ac enrichment indicates that they accumulate during S phase and mitosis and are retained at regulatory sequences until at least G1 phase and after cessation of expression of the cognate genes. We integrated our ChIPseq data with existing datasets to show that in schizont stages H3K18ac, H3K27ac and Pf H2A.Z colocalise with the transcription factor PfAP2-I and the bromodomain protein PfBDP1 and are enriched at stably positioned nucleosomes within regions of exposed DNA at active transcriptional start sites. Using transient transfections we showed that sequences enriched with colocalised H3K18ac, H3K27ac and Pf H2A.Z possess promoter activity in schizont stages, but no enhancer-like activity.
Conclusions: The dynamic H3 acetylations define P. falciparum regulatory sequences and contribute to gene activation. These findings expand the knowledge of the chromatin landscape that regulates gene expression in P. falciparum.
Keywords: Gene regulation; Histone modifications; Plasmodium falciparum.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Expression of P. falciparum var genes involves exchange of the histone variant H2A.Z at the promoter.PLoS Pathog. 2011 Feb;7(2):e1001292. doi: 10.1371/journal.ppat.1001292. Epub 2011 Feb 17. PLoS Pathog. 2011. PMID: 21379342 Free PMC article.
-
H2A.Z and H2B.Z double-variant nucleosomes define intergenic regions and dynamically occupy var gene promoters in the malaria parasite Plasmodium falciparum.Mol Microbiol. 2013 Mar;87(6):1167-82. doi: 10.1111/mmi.12154. Epub 2013 Feb 4. Mol Microbiol. 2013. PMID: 23373537
-
Actin-related protein Arp4 regulates euchromatic gene expression and development through H2A.Z deposition in blood-stage Plasmodium falciparum.Parasit Vectors. 2020 Jun 17;13(1):314. doi: 10.1186/s13071-020-04139-6. Parasit Vectors. 2020. PMID: 32552779 Free PMC article.
-
Chromatin-mediated epigenetic regulation in the malaria parasite Plasmodium falciparum.Eukaryot Cell. 2010 Aug;9(8):1138-49. doi: 10.1128/EC.00036-10. Epub 2010 May 7. Eukaryot Cell. 2010. PMID: 20453074 Free PMC article. Review.
-
Epigenetic Players of Chromatin Structure Regulation in Plasmodium falciparum.Chembiochem. 2019 May 15;20(10):1225-1230. doi: 10.1002/cbic.201800718. Epub 2019 Mar 4. Chembiochem. 2019. PMID: 30632244 Review.
Cited by
-
Genome-wide localization of histone variants in Toxoplasma gondii implicates variant exchange in stage-specific gene expression.BMC Genomics. 2022 Feb 14;23(1):128. doi: 10.1186/s12864-022-08338-6. BMC Genomics. 2022. PMID: 35164683 Free PMC article.
-
The genetic landscape of origins of replication in P. falciparum.Nucleic Acids Res. 2024 Jan 25;52(2):660-676. doi: 10.1093/nar/gkad1103. Nucleic Acids Res. 2024. PMID: 38038269 Free PMC article.
-
Plasmodium falciparum Development from Gametocyte to Oocyst: Insight from Functional Studies.Microorganisms. 2023 Jul 31;11(8):1966. doi: 10.3390/microorganisms11081966. Microorganisms. 2023. PMID: 37630530 Free PMC article. Review.
-
Distinct Histone Post-translational Modifications during Plasmodium falciparum Gametocyte Development.J Proteome Res. 2022 Aug 5;21(8):1857-1867. doi: 10.1021/acs.jproteome.2c00108. Epub 2022 Jun 30. J Proteome Res. 2022. PMID: 35772009 Free PMC article.
-
Plasmodium 6-Cysteine Proteins: Functional Diversity, Transmission-Blocking Antibodies and Structural Scaffolds.Front Cell Infect Microbiol. 2022 Jul 8;12:945924. doi: 10.3389/fcimb.2022.945924. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35899047 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

