A machine learning framework to determine geolocations from metagenomic profiling
- PMID: 33225966
- PMCID: PMC7682025
- DOI: 10.1186/s13062-020-00278-z
A machine learning framework to determine geolocations from metagenomic profiling
Abstract
Background: Studies on metagenomic data of environmental microbial samples found that microbial communities seem to be geolocation-specific, and the microbiome abundance profile can be a differentiating feature to identify samples' geolocations. In this paper, we present a machine learning framework to determine the geolocations from metagenomics profiling of microbial samples.
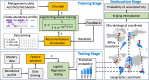
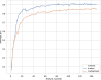
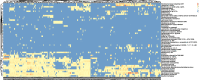
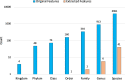
Results: Our method was applied to the multi-source microbiome data from MetaSUB (The Metagenomics and Metadesign of Subways and Urban Biomes) International Consortium for the CAMDA 2019 Metagenomic Forensics Challenge (the Challenge). The goal of the Challenge is to predict the geographical origins of mystery samples by constructing microbiome fingerprints.First, we extracted features from metagenomic abundance profiles. We then randomly split the training data into training and validation sets and trained the prediction models on the training set. Prediction performance was evaluated on the validation set. By using logistic regression with L2 normalization, the prediction accuracy of the model reaches 86%, averaged over 100 random splits of training and validation datasets.The testing data consists of samples from cities that do not occur in the training data. To predict the "mystery" cities that are not sampled before for the testing data, we first defined biological coordinates for sampled cities based on the similarity of microbial samples from them. Then we performed affine transform on the map such that the distance between cities measures their biological difference rather than geographical distance. After that, we derived the probabilities of a given testing sample from unsampled cities based on its predicted probabilities on sampled cities using Kriging interpolation. Results show that this method can successfully assign high probabilities to the true cities-of-origin of testing samples.
Conclusion: Our framework shows good performance in predicting the geographic origin of metagenomic samples for cities where training data are available. Furthermore, we demonstrate the potential of the proposed method to predict metagenomic samples' geolocations for samples from locations that are not in the training dataset.
Keywords: Abundance profiling; Affine transform; Binning; Kriging interpolation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Application of machine learning techniques for creating urban microbial fingerprints.Biol Direct. 2019 Aug 16;14(1):13. doi: 10.1186/s13062-019-0245-x. Biol Direct. 2019. PMID: 31420049 Free PMC article.
-
Massive metagenomic data analysis using abundance-based machine learning.Biol Direct. 2019 Aug 1;14(1):12. doi: 10.1186/s13062-019-0242-0. Biol Direct. 2019. PMID: 31370905 Free PMC article.
-
Systematic evaluation of supervised machine learning for sample origin prediction using metagenomic sequencing data.Biol Direct. 2020 Dec 10;15(1):29. doi: 10.1186/s13062-020-00287-y. Biol Direct. 2020. PMID: 33302990 Free PMC article.
-
A review of neural networks for metagenomic binning.Brief Bioinform. 2025 Mar 4;26(2):bbaf065. doi: 10.1093/bib/bbaf065. Brief Bioinform. 2025. PMID: 40131312 Free PMC article. Review.
-
Where environmental microbiome meets its host: Subway and passenger microbiome relationships.Mol Ecol. 2023 May;32(10):2602-2618. doi: 10.1111/mec.16440. Epub 2022 Apr 4. Mol Ecol. 2023. PMID: 35318755 Review.
Cited by
-
Artificial intelligence in forensic medicine and forensic dentistry.J Forensic Odontostomatol. 2023 Aug 27;41(2):30-41. J Forensic Odontostomatol. 2023. PMID: 37634174 Free PMC article. Review.
-
Advances in machine learning-based bacteria analysis for forensic identification: identity, ethnicity, and site of occurrence.Front Microbiol. 2023 Dec 21;14:1332857. doi: 10.3389/fmicb.2023.1332857. eCollection 2023. Front Microbiol. 2023. PMID: 38179452 Free PMC article. Review.
-
Evolution of Diagnostic and Forensic Microbiology in the Era of Artificial Intelligence.Cureus. 2023 Sep 21;15(9):e45738. doi: 10.7759/cureus.45738. eCollection 2023 Sep. Cureus. 2023. PMID: 37872929 Free PMC article. Review.
-
Advances in microbial metagenomics and artificial intelligence analysis in forensic identification.Front Microbiol. 2022 Nov 15;13:1046733. doi: 10.3389/fmicb.2022.1046733. eCollection 2022. Front Microbiol. 2022. PMID: 36458190 Free PMC article. Review.
-
A Comprehensive Insight of Current and Future Challenges in Large-Scale Soil Microbiome Analyses.Microb Ecol. 2023 Jul;86(1):75-85. doi: 10.1007/s00248-022-02060-2. Epub 2022 Jun 23. Microb Ecol. 2023. PMID: 35739325 Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

