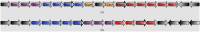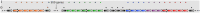Casboundary: automated definition of integral Cas cassettes
- PMID: 33226067
- PMCID: PMC8208735
- DOI: 10.1093/bioinformatics/btaa984
Casboundary: automated definition of integral Cas cassettes
Abstract
Motivation: CRISPR-Cas are important systems found in most archaeal and many bacterial genomes, providing adaptive immunity against mobile genetic elements in prokaryotes. The CRISPR-Cas systems are encoded by a set of consecutive cas genes, here termed cassette. The identification of cassette boundaries is key for finding cassettes in CRISPR research field. This is often carried out by using Hidden Markov Models and manual annotation. In this article, we propose the first method able to automatically define the cassette boundaries. In addition, we present a Cas-type predictive model used by the method to assign each gene located in the region defined by a cassette's boundaries a Cas label from a set of pre-defined Cas types. Furthermore, the proposed method can detect potentially new cas genes and decompose a cassette into its modules.
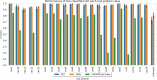
Results: We evaluate the predictive performance of our proposed method on data collected from the two most recent CRISPR classification studies. In our experiments, we obtain an average similarity of 0.86 between the predicted and expected cassettes. Besides, we achieve F-scores above 0.9 for the classification of cas genes of known types and 0.73 for the unknown ones. Finally, we conduct two additional study cases, where we investigate the occurrence of potentially new cas genes and the occurrence of module exchange between different genomes.
Availability and implementation: https://github.com/BackofenLab/Casboundary.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press.
Figures