Targeting SARS-CoV-2 main protease: structure based virtual screening, in silico ADMET studies and molecular dynamics simulation for identification of potential inhibitors
- PMID: 33226303
- PMCID: PMC7754935
- DOI: 10.1080/07391102.2020.1848636
Targeting SARS-CoV-2 main protease: structure based virtual screening, in silico ADMET studies and molecular dynamics simulation for identification of potential inhibitors
Abstract
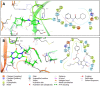
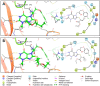
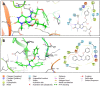
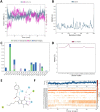
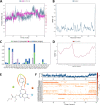
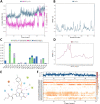
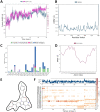
COVID-19 pandemic has created a healthcare crisis across the world and has put human life under life-threatening circumstances. The recent discovery of the crystallized structure of the main protease (Mpro) from SARS-CoV-2 has provided an opportunity for utilizing computational tools as an effective method for drug discovery. Targeting viral replication has remained an effective strategy for drug development. Mpro of SARS-COV-2 is the key protein in viral replication as it is involved in the processing of polyproteins to various structural and nonstructural proteins. Thus, Mpro represents a key target for the inhibition of viral replication specifically for SARS-CoV-2. We have used a virtual screening strategy by targeting Mpro against a library of commercially available compounds to identify potential inhibitors. After initial identification of hits by molecular docking-based virtual screening further MM/GBSA, predictive ADME analysis, and molecular dynamics simulation were performed. The virtual screening resulted in the identification of twenty-five top scoring structurally diverse hits that have free energy of binding (ΔG) values in the range of -26-06 (for compound AO-854/10413043) to -59.81 Kcal/mol (for compound 329/06315047). Moreover, the top-scoring hits have favorable AMDE properties as calculated using in silico algorithms. Additionally, the molecular dynamics simulation revealed the stable nature of protein-ligand interaction and provided information about the amino acid residues involved in binding. Overall, this study led to the identification of potential SARS-CoV-2 Mpro hit compounds with favorable pharmacokinetic properties. We believe that the outcome of this study can help to develop novel Mpro inhibitors to tackle this pandemic.Communicated by Ramaswamy H. Sarma.
Keywords: SARS-COV-2; Virtual screening; anti-viral; hit identification; in silico ADME; main protease; molecular dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Isatin-based virtual high throughput screening, molecular docking, DFT, QM/MM, MD and MM-PBSA study of novel inhibitors of SARS-CoV-2 main protease.J Biomol Struct Dyn. 2022 Oct;40(17):7852-7867. doi: 10.1080/07391102.2021.1904003. Epub 2021 Mar 25. J Biomol Struct Dyn. 2022. PMID: 33764269
-
In silico identification of potential inhibitors of key SARS-CoV-2 3CL hydrolase (Mpro) via molecular docking, MMGBSA predictive binding energy calculations, and molecular dynamics simulation.PLoS One. 2020 Jul 24;15(7):e0235030. doi: 10.1371/journal.pone.0235030. eCollection 2020. PLoS One. 2020. PMID: 32706783 Free PMC article.
-
Identification of natural inhibitors against Mpro of SARS-CoV-2 by molecular docking, molecular dynamics simulation, and MM/PBSA methods.J Biomol Struct Dyn. 2022 Apr;40(6):2757-2768. doi: 10.1080/07391102.2020.1842806. Epub 2020 Nov 4. J Biomol Struct Dyn. 2022. PMID: 33143552 Free PMC article.
-
Covalent small-molecule inhibitors of SARS-CoV-2 Mpro: Insights into their design, classification, biological activity, and binding interactions.Eur J Med Chem. 2024 Nov 5;277:116704. doi: 10.1016/j.ejmech.2024.116704. Epub 2024 Aug 8. Eur J Med Chem. 2024. PMID: 39121741 Review.
-
Potential Resistance of SARS-CoV-2 Main Protease (Mpro) against Protease Inhibitors: Lessons Learned from HIV-1 Protease.Int J Mol Sci. 2022 Mar 23;23(7):3507. doi: 10.3390/ijms23073507. Int J Mol Sci. 2022. PMID: 35408866 Free PMC article. Review.
Cited by
-
In silico evaluation of anti-colorectal cancer inhibitors by Resveratrol derivatives targeting Armadillo repeats domain of APC: molecular docking and molecular dynamics simulation.Front Oncol. 2024 Apr 30;14:1360745. doi: 10.3389/fonc.2024.1360745. eCollection 2024. Front Oncol. 2024. PMID: 38746675 Free PMC article.
-
Towards the discovery of potential RdRp inhibitors for the treatment of COVID-19: structure guided virtual screening, computational ADME and molecular dynamics study.Struct Chem. 2022;33(5):1569-1583. doi: 10.1007/s11224-022-01976-2. Epub 2022 Jun 2. Struct Chem. 2022. PMID: 35669792 Free PMC article.
-
Urea-thiazole/benzothiazole hybrids with a triazole linker: synthesis, antimicrobial potential, pharmacokinetic profile and in silico mechanistic studies.Mol Divers. 2022 Oct;26(5):2375-2391. doi: 10.1007/s11030-021-10336-x. Epub 2021 Oct 20. Mol Divers. 2022. PMID: 34671895
-
Novel Drug Design for Treatment of COVID-19: A Systematic Review of Preclinical Studies.Can J Infect Dis Med Microbiol. 2022 Sep 25;2022:2044282. doi: 10.1155/2022/2044282. eCollection 2022. Can J Infect Dis Med Microbiol. 2022. PMID: 36199815 Free PMC article. Review.
-
Identification of a novel inhibitor of SARS-CoV-2 3CL-PRO through virtual screening and molecular dynamics simulation.PeerJ. 2021 Apr 13;9:e11261. doi: 10.7717/peerj.11261. eCollection 2021. PeerJ. 2021. PMID: 33954055 Free PMC article.
References
-
- Adeshina, Y. O., Deeds, E. J., & Karanicolas, J. (2020). Machine learning classification can reduce false positives in structure-based virtual screening. Proceedings of the National Academy of Sciences of the United States of America, 117(31), 18477–18488. 10.1073/pnas.2000585117 - DOI - PMC - PubMed
-
- Alavijeh, M. S., & Palmer, A. M. (2004). The pivotal role of drug metabolism and pharmacokinetics in the discovery and development of new medicines. IDrugs : The Investigational Drugs Journal, 7(8), 755–763. - PubMed
-
- Asinex . (2020). http://www.asinex.com/libraries-html/libraries_gold_platinum-html/
-
- Bowers, K. J. (2006). Scalable algorithms for molecular dynamics simulations on commodity clusters. SC '06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing (pp. 43–43), November 11–17. Institute of Electrical and Electronics Engineers (IEEE). 10.1109/SC.2006.54 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
