Clinical exome sequencing is a powerful tool in the diagnostic flow of monogenic kidney diseases: an Italian experience
- PMID: 33226606
- PMCID: PMC8494711
- DOI: 10.1007/s40620-020-00898-8
Clinical exome sequencing is a powerful tool in the diagnostic flow of monogenic kidney diseases: an Italian experience
Abstract
Background: A considerable minority of patients on waiting lists for kidney transplantation either have no diagnosis (and fall into the subset of undiagnosed cases) because kidney biopsy was not performed or histological findings were non-specific, or do not fall into any well-defined clinical category. Some of these patients might be affected by a previously unrecognised monogenic disease.
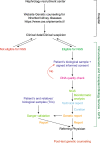
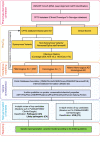
Methods: Through a multidisciplinary cooperative effort, we built an analytical pipeline to identify patients with chronic kidney disease (CKD) with a clinical suspicion of a monogenic condition or without a well-defined diagnosis. Following the stringent phenotypical and clinical characterization required by the flowchart, candidates meeting these criteria were further investigated by clinical exome sequencing followed by in silico analysis of 225 kidney-disease-related genes.
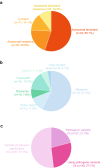
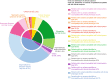
Results: By using an ad hoc web-based platform, we enrolled 160 patients from 13 different Nephrology and Genetics Units located across the Piedmont region over 15 months. A preliminary "remote" evaluation based on well-defined inclusion criteria allowed us to define eligibility for NGS analysis. Among the 138 recruited patients, 52 (37.7%) were children and 86 (62.3%) were adults. Up to 48% of them had a positive family history for kidney disease. Overall, applying this workflow led to the identification of genetic variants potentially explaining the phenotype in 78 (56.5%) cases.
Conclusions: These results underline the importance of clinical exome sequencing as a versatile and highly useful, non-invasive tool for genetic diagnosis of kidney diseases. Identifying patients who can benefit from targeted therapies, and improving the management of organ transplantation are further expected applications.
Keywords: Chronic kidney failure; Next-generation sequencing; Renal monogenic disease; Transplantation.
© 2020. The Author(s).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Connaughton DM, Bukhari S, Conlon P, Cassidy E, O'Toole M, Mohamad M, Flanagan J, Butler T, O'Leary A, Wong L, O'Regan J, Moran S, O'Kelly P, Logan V, Griffin B, Griffin M, Lavin P, Little MA, Conlon P. The Irish kidney gene project-prevalence of family history in patients with kidney disease in Ireland. Nephron. 2015;130(4):293–301. doi: 10.1159/000436983. - DOI - PubMed

