Genomic surveillance of SARS-CoV-2 in Thailand reveals mixed imported populations, a local lineage expansion and a virus with truncated ORF7a
- PMID: 33227343
- PMCID: PMC7679658
- DOI: 10.1016/j.virusres.2020.198233
Genomic surveillance of SARS-CoV-2 in Thailand reveals mixed imported populations, a local lineage expansion and a virus with truncated ORF7a
Abstract
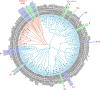
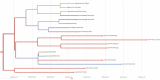
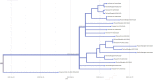
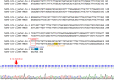
Coronavirus Disease 2019 (COVID-19) is a global public health threat. Genomic surveillance of SARS-CoV-2 was implemented in March of 2020 at a major diagnostic hub in Bangkok, Thailand. Several virus lineages supposedly originated in many countries were found, and a Thai-specific lineage, designated A/Thai-1, has expanded to be predominant in Thailand. A virus sample in the SARS-CoV-2 A/Thai-1 lineage contains a frame-shift deletion at ORF7a, encoding a putative host antagonizing factor of the virus.
Keywords: COVID-19; Genomic surveillance; SARS-CoV-2; Thailand.
Copyright © 2020. Published by Elsevier B.V.
Conflict of interest statement
None.
Figures




References
-
- Giovanetti M. Genomic and epidemiological surveillance of zika virus in the Amazon region. Cell Rep. 2020;30(7):2275–2283. e7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

