Connection between the Gut Microbiome, Systemic Inflammation, Gut Permeability and FOXP3 Expression in Patients with Primary Sjögren's Syndrome
- PMID: 33228011
- PMCID: PMC7699261
- DOI: 10.3390/ijms21228733
Connection between the Gut Microbiome, Systemic Inflammation, Gut Permeability and FOXP3 Expression in Patients with Primary Sjögren's Syndrome
Abstract
The aims of this study were to explore intestinal microbial composition and functionality in primary Sjögren's syndrome (pSS) and to relate these findings to inflammation, permeability and the transcription factor Forkhead box protein P3 (FOXP3) gene expression in peripheral blood. The study included 19 pSS patients and 19 healthy controls matched for age, sex, and body mass index. Fecal bacterial DNA was extracted and analyzed by 16S rRNA sequencing using an Ion S5 platform followed by a bioinformatics analysis using Quantitative Insights into Microbial Ecology (QIIME II) and Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt). Our data suggest that the gut microbiota of pSS patients differs at both the taxonomic and functional levels with respect to healthy controls. The gut microbiota profile of our pSS patients was characterized by a lower diversity and richness and with Bacteroidetes dominating at the phylum level. The pSS patients had less beneficial or commensal butyrate-producing bacteria and a higher proportion of opportunistic pathogens with proinflammatory activity, which may impair intestinal barrier function and therefore contribute to inflammatory processes associated with pSS by increasing the production of proinflammatory cytokines and decreasing the release of the anti-inflammatory cytokine IL-10 and the peripheral FOXP3 mRNA expression, implicated in the development and function of regulatory T cells (Treg) cells. Further studies are needed to better understand the real impact of dysbiosis on the course of pSS and to conceive preventive or therapeutic strategies to counteract microbiome-driven inflammation.
Keywords: FOXP3 expression; gut microbiota; inflammation; intestinal permeability; primary Sjögren’s syndrome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
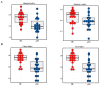

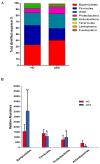
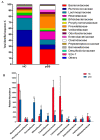
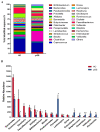

References
-
- Carsons S. A review and update of Sjögren’s syndrome: Manifestations, diagnosis, and treatment. Am. J. Manag. Care. 2001;7:S433–S443. - PubMed
MeSH terms
Substances
Grants and funding
- CPI13/00003/Miguel Servet Type II" program, ISCIII, Spain
- C-0030-2018/"Nicolas Monardes" research program of the Consejería de Salud (C-0030-2018, Junta de Andalucía, Spain
- PE-0106-2019/Predoctoral grant from Consejería de Salud y Familia, Andalucia, Spain
- FI19-00112/predoctoral grant PFIS-ISCIII, Madrid, Spain

