Proteomic Studies of the Biofilm Matrix including Outer Membrane Vesicles of Burkholderia multivorans C1576, a Strain of Clinical Importance for Cystic Fibrosis
- PMID: 33228110
- PMCID: PMC7699398
- DOI: 10.3390/microorganisms8111826
Proteomic Studies of the Biofilm Matrix including Outer Membrane Vesicles of Burkholderia multivorans C1576, a Strain of Clinical Importance for Cystic Fibrosis
Abstract
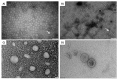
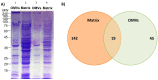
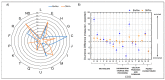
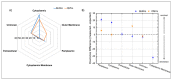
Biofilms are aggregates of microbial cells encased in a highly hydrated matrix made up of self-produced extracellular polymeric substances (EPS) which consist of polysaccharides, proteins, nucleic acids, and lipids. While biofilm matrix polysaccharides are unraveled, there is still poor knowledge about the identity and function of matrix-associated proteins. With this work, we performed a comprehensive proteomic approach to disclose the identity of proteins associated with the matrix of biofilm-growing Burkholderia multivorans C1576 reference strain, a cystic fibrosis clinical isolate. Transmission electron microscopy showed that B. multivorans C1576 also releases outer membrane vesicles (OMVs) in the biofilm matrix, as already demonstrated for other Gram-negative species. The proteomic analysis revealed that cytoplasmic and membrane-bound proteins are widely represented in the matrix, while OMVs are highly enriched in outer membrane proteins and siderophores. Our data suggest that cell lysis and OMVs production are the most important sources of proteins for the B. multivorans C1576 biofilm matrix. Of note, some of the identified proteins are lytic enzymes, siderophores, and proteins involved in reactive oxygen species (ROS) scavenging. These proteins might help B. multivorans C1576 in host tissue invasion and defense towards immune system assaults.
Keywords: Burkholderia multivorans; LC-LC/MS; biofilm; outer membrane vesicles; proteomic.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

