Plant Organellar DNA Polymerases Evolved Multifunctionality through the Acquisition of Novel Amino Acid Insertions
- PMID: 33228188
- PMCID: PMC7699545
- DOI: 10.3390/genes11111370
Plant Organellar DNA Polymerases Evolved Multifunctionality through the Acquisition of Novel Amino Acid Insertions
Abstract
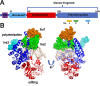
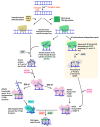
The majority of DNA polymerases (DNAPs) are specialized enzymes with specific roles in DNA replication, translesion DNA synthesis (TLS), or DNA repair. The enzymatic characteristics to perform accurate DNA replication are in apparent contradiction with TLS or DNA repair abilities. For instance, replicative DNAPs incorporate nucleotides with high fidelity and processivity, whereas TLS DNAPs are low-fidelity polymerases with distributive nucleotide incorporation. Plant organelles (mitochondria and chloroplast) are replicated by family-A DNA polymerases that are both replicative and TLS DNAPs. Furthermore, plant organellar DNA polymerases from the plant model Arabidopsis thaliana (AtPOLIs) execute repair of double-stranded breaks by microhomology-mediated end-joining and perform Base Excision Repair (BER) using lyase and strand-displacement activities. AtPOLIs harbor three unique insertions in their polymerization domain that are associated with TLS, microhomology-mediated end-joining (MMEJ), strand-displacement, and lyase activities. We postulate that AtPOLIs are able to execute those different functions through the acquisition of these novel amino acid insertions, making them multifunctional enzymes able to participate in DNA replication and DNA repair.
Keywords: DNA repair; DNA replication; chloroplast; mitochondria; plant organellar DNA polymerases.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

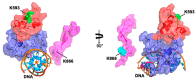

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

