Global analysis of the AP2/ERF gene family in rose (Rosa chinensis) genome unveils the role of RcERF099 in Botrytis resistance
- PMID: 33228522
- PMCID: PMC7684944
- DOI: 10.1186/s12870-020-02740-6
Global analysis of the AP2/ERF gene family in rose (Rosa chinensis) genome unveils the role of RcERF099 in Botrytis resistance
Abstract
Background: The AP2/ERFs belong to a large family of transcription factors in plants. The AP2/ERF gene family has been identified as a key player involved in both biotic and abiotic stress responses in plants, however, no comprehensive study has yet been carried out on the AP2/ERF gene family in rose (Rosa sp.), the most important ornamental crop worldwide.
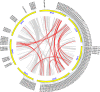
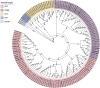
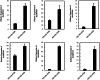
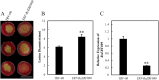
Results: The present study comprises a genome-wide analysis of the AP2/ERF family genes (RcERFs) in the rose, involving their identification, gene structure, phylogenetic relationship, chromosome localization, collinearity analysis, as well as their expression patterns. Throughout the phylogenetic analysis, a total of 131 AP2/ERF genes in the rose genome were divided into 5 subgroups. The RcERFs are distributed over all the seven chromosomes of the rose, and genome duplication may have played a key role in their duplication. Furthermore, Ka/Ks analysis indicated that the duplicated RcERF genes often undergo purification selection with limited functional differentiation. Gene expression analysis revealed that 23 RcERFs were induced by infection of the necrotrophic fungal pathogen Botrytis cinerea. Presumably, these RcERFs are candidate genes which can react to the rose's resistance against Botrytis cinerea infection. By using virus-induced gene silencing, we confirmed that RcERF099 is an important regulator involved in the B.cinerea resistance in the rose petal.
Conclusion: Overall, our results conclude the necessity for further study of the AP2/ERF gene family in rose, and promote their potential application in improving the rose when subjected to biological stress.
Keywords: AP2/ERF gene family; Botrytis cinerea; Rosa sp.; Virus-induced gene silencing.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290(3):998–1009. doi: 10.1006/bbrc.2001.6299. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 31772344/National Natural Science Foundation of China
- 31972444/National Natural Science Foundation of China
- 2019JQ645/Natural Science Foundation of Shaanxi Province
- 2452019112/Fundamental Research Funds for the Central Universities
- 2452019040/Scientific Research Startup Fund for Talents, Northwest A&F University
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

