Bile Salts Differentially Enhance Resistance of Enterohemorrhagic Escherichia coli O157:H7 to Host Defense Peptides
- PMID: 33229368
- PMCID: PMC7822141
- DOI: 10.1128/IAI.00719-20
Bile Salts Differentially Enhance Resistance of Enterohemorrhagic Escherichia coli O157:H7 to Host Defense Peptides
Abstract
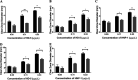
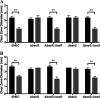
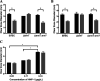
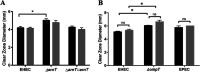
During passage through the human gastrointestinal tract, enterohemorrhagic Escherichia coli (EHEC) is exposed to membrane-damaging bile in the small intestine. We previously reported that EHEC treatment with a physiological bile salt mixture upregulates basRS, encoding a two-component system, and arnBCADTEF, encoding the aminoarabinose lipid A modification pathway (J. V. Kus, A. Gebremedhin, V. Dang, S. L. Tran, A. Serbanescu, and D. Barnett Foster, J Bacteriol 193: 4509-4515, 2011, https://doi.org/10.1128/JB.00200-11). The present study examined the effect of bile salt mix (BSM) treatment on EHEC resistance to three human gastrointestinal defense peptides-HD-5, HNP-1, and LL-37-as well as the role of basRS and arnT in the respective responses. After BSM treatment, EHEC resistance to HD-5 and HNP-1 was significantly increased in a BSM-, defensin dose-dependent manner. The resistance phenotype was dependent on both basRS and arnT However, the BSM treatment did not alter EHEC resistance to LL-37, even when the ompT gene, encoding an LL-37 cleavage protease, was disrupted. Interestingly, enteropathogenic E. coli, a related pathogen that infects the small intestine, showed a similar BSM-induced resistance phenotype. Using a model of EHEC infection in Galleria mellonella, we found significantly lower survival rates in wax moth larvae infected with BSM-treated wild-type EHEC than in those infected with a BSM-treated basS mutant, suggesting that treatment with a physiological BSM enhances virulence through a basS-mediated pathway. The results of this investigation provide persuasive evidence that bile salts typically encountered during transit through the small intestine can serve as an environmental cue for EHEC, enhancing resistance to several key host defense peptides.
Keywords: antimicrobial resistance; bile salts; defensins; enterohemorrhagic E. coli; gastrointestinal infection; host defense peptides.
Copyright © 2021 American Society for Microbiology.
Figures

Similar articles
-
Molecular basis of bile-salt- and iron-induced enterohaemorrhagic E. coli resistance to cationic antimicrobial peptides.Microbiology (Reading). 2020 Dec;166(12):1149-1159. doi: 10.1099/mic.0.000988. Microbiology (Reading). 2020. PMID: 33205745
-
Bile salts induce resistance to polymyxin in enterohemorrhagic Escherichia coli O157:H7.J Bacteriol. 2011 Sep;193(17):4509-15. doi: 10.1128/JB.00200-11. Epub 2011 Jul 1. J Bacteriol. 2011. PMID: 21725004 Free PMC article.
-
Enterohemorrhagic and enteropathogenic Escherichia coli evolved different strategies to resist antimicrobial peptides.Gut Microbes. 2012 Nov-Dec;3(6):556-61. doi: 10.4161/gmic.21656. Epub 2012 Aug 16. Gut Microbes. 2012. PMID: 22895086 Free PMC article. Review.
-
OmpT outer membrane proteases of enterohemorrhagic and enteropathogenic Escherichia coli contribute differently to the degradation of human LL-37.Infect Immun. 2012 Feb;80(2):483-92. doi: 10.1128/IAI.05674-11. Epub 2011 Dec 5. Infect Immun. 2012. PMID: 22144482 Free PMC article.
-
Phytochemicals Controlling Enterohemorrhagic Escherichia coli (EHEC) Virulence-Current Knowledge of Their Mechanisms of Action.Int J Mol Sci. 2025 Jan 4;26(1):381. doi: 10.3390/ijms26010381. Int J Mol Sci. 2025. PMID: 39796236 Free PMC article. Review.
Cited by
-
Enhanced antibacterial activity of acid treated MgO nanoparticles on Escherichia coli.RSC Adv. 2021 Nov 29;11(60):38202-38207. doi: 10.1039/d1ra06221b. eCollection 2021 Nov 23. RSC Adv. 2021. PMID: 35498104 Free PMC article.
-
The gut-liver axis in hepatobiliary diseases.Inflamm Regen. 2024 Jan 8;44(1):2. doi: 10.1186/s41232-023-00315-0. Inflamm Regen. 2024. PMID: 38191517 Free PMC article. Review.
-
Checkpoints That Regulate Balanced Biosynthesis of Lipopolysaccharide and Its Essentiality in Escherichia coli.Int J Mol Sci. 2021 Dec 24;23(1):189. doi: 10.3390/ijms23010189. Int J Mol Sci. 2021. PMID: 35008618 Free PMC article. Review.
-
Impact of Western Diet on Enterohemorrhagic Escherichia coli Colonization in the Human In Vitro Mucosal Artificial Colon as Mediated by Gut Microbiota.Nutrients. 2024 Jun 27;16(13):2046. doi: 10.3390/nu16132046. Nutrients. 2024. PMID: 38999794 Free PMC article.
References
-
- Karpman D, Ståhl A. 2014. Enterohemorrhagic Escherichia coli pathogenesis and the host response, p 403–417. In Sperandio V, Hovde CJ (ed), Enterohemorrhagic Escherichia coli and other Shiga toxin-producing E. coli. ASM Press, Washington, DC. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

