The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles
- PMID: 33229438
- PMCID: PMC7833635
- DOI: 10.1074/jbc.RA120.016175
The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles
Abstract
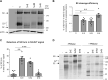
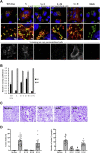
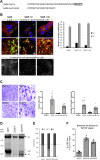
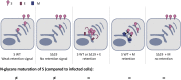
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a β-coronavirus, is the causative agent of the COVID-19 pandemic. Like for other coronaviruses, its particles are composed of four structural proteins: spike (S), envelope (E), membrane (M), and nucleoprotein (N) proteins. The involvement of each of these proteins and their interactions are critical for assembly and production of β-coronavirus particles. Here, we sought to characterize the interplay of SARS-CoV-2 structural proteins during the viral assembly process. By combining biochemical and imaging assays in infected versus transfected cells, we show that E and M regulate intracellular trafficking of S as well as its intracellular processing. Indeed, the imaging data reveal that S is relocalized at endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) or Golgi compartments upon coexpression of E or M, as observed in SARS-CoV-2-infected cells, which prevents syncytia formation. We show that a C-terminal retrieval motif in the cytoplasmic tail of S is required for its M-mediated retention in the ERGIC, whereas E induces S retention by modulating the cell secretory pathway. We also highlight that E and M induce a specific maturation of N-glycosylation of S, independently of the regulation of its localization, with a profile that is observed both in infected cells and in purified viral particles. Finally, we show that E, M, and N are required for optimal production of virus-like-particles. Altogether, these results highlight how E and M proteins may influence the properties of S proteins and promote the assembly of SARS-CoV-2 viral particles.
Keywords: COVID-19; SARS-CoV; glycoprotein; infectious disease; secretion; viral protein; virus assembly.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- WHO. Coronavirus Disease (COVID-2019) situation reports.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

