ClpX Is Essential and Activated by Single-Strand DNA Binding Protein in Mycobacteria
- PMID: 33229461
- PMCID: PMC7847540
- DOI: 10.1128/JB.00608-20
ClpX Is Essential and Activated by Single-Strand DNA Binding Protein in Mycobacteria
Abstract
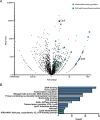
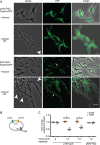
The ClpP1P2 proteolytic complex is essential in Mycobacterium tuberculosis Proteolysis by ClpP1P2 requires an associated ATPase, either ClpX or ClpC1. Here, we sought to define the unique contributions of the ClpX ATPase to mycobacterial growth. We formally demonstrated that ClpX is essential for mycobacterial growth, and to understand its essential functions, we identified ClpX-His-interacting proteins by pulldown and tandem mass spectrometry. We found an unexpected association between ClpX and proteins involved in DNA replication, and we confirm a physical association between ClpX and the essential DNA maintenance protein single-stranded-DNA binding protein (SSB). Purified SSB is not degraded by ClpXP1P2; instead, SSB enhances ATP hydrolysis by ClpX and degradation of the model substrate GFP-SsrA by ClpXP1P2. This activation of ClpX is mediated by the C-terminal tail of SSB, which had been implicated in the activation of other ATPases associated with DNA replication. Consistent with the predicted interactions, depletion of clpX transcript perturbs DNA replication. These data reveal that ClpX participates in DNA replication and identify the first activator of ClpX in mycobacteria.IMPORTANCE Tuberculosis, caused by Mycobacterium tuberculosis, imposes a major global health burden, surpassing HIV and malaria in annual deaths. The ClpP1P2 proteolytic complex and its cofactor ClpX are attractive drug targets, but their precise cellular functions are unclear. This work confirms ClpX's essentiality and describes a novel interaction between ClpX and SSB, a component of the DNA replication machinery. Further, we demonstrate that a loss of ClpX is sufficient to interrupt DNA replication, suggesting that the ClpX-SSB complex may play a role in DNA replication in mycobacteria.
Keywords: DNA replication; cell cycle; mycobacteria; protein degradation.
Copyright © 2021 Kester et al.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

