High-resolution mouse subventricular zone stem-cell niche transcriptome reveals features of lineage, anatomy, and aging
- PMID: 33229571
- PMCID: PMC7733854
- DOI: 10.1073/pnas.2014389117
High-resolution mouse subventricular zone stem-cell niche transcriptome reveals features of lineage, anatomy, and aging
Abstract
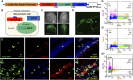
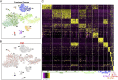
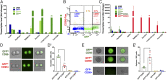
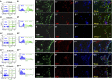
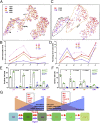
Adult neural stem cells (NSC) serve as a reservoir for brain plasticity and origin for certain gliomas. Lineage tracing and genomic approaches have portrayed complex underlying heterogeneity within the major anatomical location for NSC, the subventricular zone (SVZ). To gain a comprehensive profile of NSC heterogeneity, we utilized a well-validated stem/progenitor-specific reporter transgene in concert with single-cell RNA sequencing to achieve unbiased analysis of SVZ cells from infancy to advanced age. The magnitude and high specificity of the resulting transcriptional datasets allow precise identification of the varied cell types embedded in the SVZ including specialized parenchymal cells (neurons, glia, microglia) and noncentral nervous system cells (endothelial, immune). Initial mining of the data delineates four quiescent NSC and three progenitor-cell subpopulations formed in a linear progression. Further evidence indicates that distinct stem and progenitor populations reside in different regions of the SVZ. As stem/progenitor populations progress from neonatal to advanced age, they acquire a deficiency in transition from quiescence to proliferation. Further data mining identifies stage-specific biological processes, transcription factor networks, and cell-surface markers for investigation of cellular identities, lineage relationships, and key regulatory pathways in adult NSC maintenance and neurogenesis.
Keywords: aging; neural stem cell; single-cell RNA sequencing; subventricular zone; transcriptome.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Altman J., Das G. D., Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 124, 319–335 (1965). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

