ETNK1 mutations induce a mutator phenotype that can be reverted with phosphoethanolamine
- PMID: 33230096
- PMCID: PMC7684297
- DOI: 10.1038/s41467-020-19721-w
ETNK1 mutations induce a mutator phenotype that can be reverted with phosphoethanolamine
Abstract
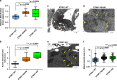
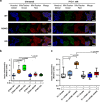
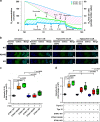
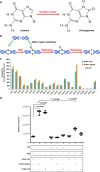
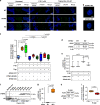
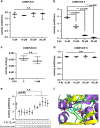
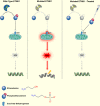
Recurrent somatic mutations in ETNK1 (Ethanolamine-Kinase-1) were identified in several myeloid malignancies and are responsible for a reduced enzymatic activity. Here, we demonstrate in primary leukemic cells and in cell lines that mutated ETNK1 causes a significant increase in mitochondrial activity, ROS production, and Histone H2AX phosphorylation, ultimately driving the increased accumulation of new mutations. We also show that phosphoethanolamine, the metabolic product of ETNK1, negatively controls mitochondrial activity through a direct competition with succinate at mitochondrial complex II. Hence, reduced intracellular phosphoethanolamine causes mitochondria hyperactivation, ROS production, and DNA damage. Treatment with phosphoethanolamine is able to counteract complex II hyperactivation and to restore a normal phenotype.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Kennedy EP, Weiss SB. The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 1956;222:193–214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

