Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes
- PMID: 33230107
- PMCID: PMC7683540
- DOI: 10.1038/s41467-020-19722-9
Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes
Abstract
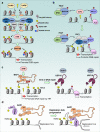
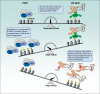
Histone posttranslational modifications are key regulators of chromatin-associated processes including gene expression, DNA replication and DNA repair. Monoubiquitinated histone H2A, H2Aub (K118 in Drosophila or K119 in vertebrates) is catalyzed by the Polycomb group (PcG) repressive complex 1 (PRC1) and reversed by the PcG-repressive deubiquitinase (PR-DUB)/BAP1 complex. Here we critically assess the current knowledge regarding H2Aub deposition and removal, its crosstalk with PcG repressive complex 2 (PRC2)-mediated histone H3K27 methylation, and the recent attempts toward discovering its readers and solving its enigmatic functions. We also discuss mounting evidence of the involvement of H2A ubiquitination in human pathologies including cancer, while highlighting some knowledge gaps that remain to be addressed.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

