ClassifyCNV: a tool for clinical annotation of copy-number variants
- PMID: 33230148
- PMCID: PMC7683568
- DOI: 10.1038/s41598-020-76425-3
ClassifyCNV: a tool for clinical annotation of copy-number variants
Abstract
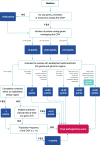
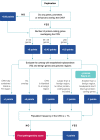
Copy-number variants (CNVs) are an important part of human genetic variation. They can be benign or can play a role in human disease by creating dosage imbalances and disrupting genes and regulatory elements. Accurate identification and clinical annotation of CNVs is essential, however, manual evaluation of individual CNVs by clinicians is challenging on a large scale. Here, we present ClassifyCNV, an easy-to-use tool that implements the 2019 ACMG classification guidelines to assess CNV pathogenicity. ClassifyCNV uses genomic coordinates and CNV type as input and reports a clinical classification for each variant, a classification score breakdown, and a list of genes of potential importance for variant interpretation. We validate ClassifyCNV's performance using a set of known clinical CNVs and a set of manually evaluated variants. ClassifyCNV matches the pathogenicity category for 81% of manually evaluated variants with the significance of the remaining pathogenic and benign variants automatically determined as uncertain, requiring a further evaluation by a clinician. ClassifyCNV facilitates the implementation of the latest ACMG guidelines in high-throughput CNV analysis, is suitable for integration into NGS analysis pipelines, and can decrease time to diagnosis. The tool is available at https://github.com/Genotek/ClassifyCNV .
Conflict of interest statement
T.A.G. and V.V.I. are employees of Genotek Ltd. The authors declare no other competing interests.
Figures
References
-
- Riggs ER, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) Genet. Med. 2020;22:245–257. doi: 10.1038/s41436-019-0686-8. - DOI - PMC - PubMed