Surveying biomolecular frustration at atomic resolution
- PMID: 33230150
- PMCID: PMC7683549
- DOI: 10.1038/s41467-020-19560-9
Surveying biomolecular frustration at atomic resolution
Abstract
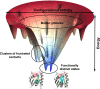
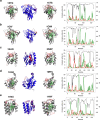
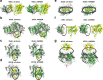
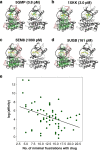
To function, biomolecules require sufficient specificity of interaction as well as stability to live in the cell while still being able to move. Thermodynamic stability of only a limited number of specific structures is important so as to prevent promiscuous interactions. The individual interactions in proteins, therefore, have evolved collectively to give funneled minimally frustrated landscapes but some strategic parts of biomolecular sequences located at specific sites in the structure have been selected to be frustrated in order to allow both motion and interaction with partners. We describe a framework efficiently to quantify and localize biomolecular frustration at atomic resolution by examining the statistics of the energy changes that occur when the local environment of a site is changed. The location of patches of highly frustrated interactions correlates with key biological locations needed for physiological function. At atomic resolution, it becomes possible to extend frustration analysis to protein-ligand complexes. At this resolution one sees that drug specificity is correlated with there being a minimally frustrated binding pocket leading to a funneled binding landscape. Atomistic frustration analysis provides a route for screening for more specific compounds for drug discovery.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

