Weak genetic signal for phenotypic integration implicates developmental processes as major regulators of trait covariation
- PMID: 33231336
- PMCID: PMC8811731
- DOI: 10.1111/mec.15748
Weak genetic signal for phenotypic integration implicates developmental processes as major regulators of trait covariation
Abstract
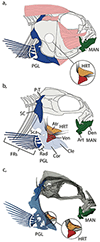
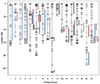
Phenotypic integration is an important metric that describes the degree of covariation among traits in a population, and is hypothesized to arise due to selection for shared functional processes. Our ability to identify the genetic and/or developmental underpinnings of integration is marred by temporally overlapping cell-, tissue- and structure-level processes that serve to continually 'overwrite' the structure of covariation among traits through ontogeny. Here, we examine whether traits that are integrated at the phenotypic level also exhibit a shared genetic basis (e.g. pleiotropy). We micro-CT scanned two hard tissue traits, and two soft tissue traits (mandible, pectoral girdle, atrium and ventricle, respectively) from an F5 hybrid population of Lake Malawi cichlids, and used geometric morphometrics to extract 3D shape information from each trait. Given the large degree of asymmetric variation that may reflect developmental instability, we separated symmetric from asymmetric components of shape variation. We then performed quantitative trait loci (QTL) analysis to determine the degree of genetic overlap between shapes. While we found ubiquitous associations among traits at the phenotypic level, except for a handful of notable exceptions, our QTL analysis revealed few overlapping genetic regions. Taken together, this indicates developmental interactions can play a large role in determining the degree of phenotypic integration among traits, and likely obfuscate the genotype to phenotype map, limiting our ability to gain a comprehensive picture of the genetic contributors responsible for phenotypic divergence.
Keywords: bone; development; geometric morphometrics; heart; phenotypic integration.
© 2020 John Wiley & Sons Ltd.
Figures






References
-
- Adams DC, and Collyer ML. 2016. On the comparison of the strength of morphological integration across morphometric datasets. Evolution (N. Y). 70:2623–2631. John Wiley & Sons, Ltd (10.1111). - PubMed
-
- Adams DC, Collyer ML, Otarola-Castillo E, and Sherratt E. 2017. Geomorph: Software for geometric morphometric analyses. CRAN.
-
- Adhikari K, Fuentes-Guajardo M, Quinto-Sánchez M, Mendoza-Revilla J, Camilo Chacón-Duque J, Acuña-Alonzo V, Jaramillo C, Arias W, Lozano RB, Pérez GM, Gómez-Valdés J, Villamil-Ramírez H, Hunemeier T, Ramallo V, Silva de Cerqueira CC, Hurtado M, Villegas V, Granja V, Gallo C, Poletti G, Schuler-Faccini L, Salzano FM, Bortolini M-C, Canizales-Quinteros S, Cheeseman M, Rosique J, Bedoya G, Rothhammer F, Headon D, González-José R, Balding D, and Ruiz-Linares A. 2016. A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat. Commun. 7:11616. - PMC - PubMed
-
- Albertson RC, and Kocher TD. 2006. Genetic and developmental basis of cichlid trophic diversity. Heredity (Edinb). 97:211–221. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

