Long non-coding RNA 01126 promotes periodontitis pathogenesis of human periodontal ligament cells via miR-518a-5p/HIF-1α/MAPK pathway
- PMID: 33231338
- PMCID: PMC7791173
- DOI: 10.1111/cpr.12957
Long non-coding RNA 01126 promotes periodontitis pathogenesis of human periodontal ligament cells via miR-518a-5p/HIF-1α/MAPK pathway
Abstract
Background: Periodontitis is a prevalent oral inflammatory disease, which can cause periodontal ligament to a local hypoxia environment. However, the mechanism of hypoxia associated long non-coding RNAs (lncRNAs) involved in periodontitis is still largely unknown.
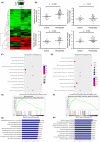
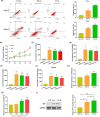
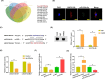
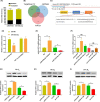
Methods: Microarray was performed to detect the expression patterns of lncRNAs in 3 pairs of gingival tissues from patients with periodontitis and healthy controls. The expression of lncRNA 01126 (LINC01126), miR-518a-5p and hypoxia-inducible factor-1α (HIF-1α) in periodontal tissues and in human periodontal ligament cells (hPDLCs) under hypoxia was measured by quantitative real-time polymerase chain reaction or western blot. Fluorescence in situ hybridization and cell fraction assay were performed to determine the subcellular localization of LINC01126 and miR-518a-5p. Overexpression or knockdown of LINC01126 or HIF-1α was used to confirm their biological roles in hPDLCs. MTT assays were performed to evaluate hPDLCs proliferation ability. Flow cytometry was used to detect apoptosis. ELISA was used to measure the expression levels of interleukin (IL)-1β, IL-6, IL-8 and TNF-α. Dual-luciferase reporter assays were performed to assess the binding of miR-518a-5p to LINC01126 and HIF-1α. RNA immunoprecipitation assay was used to identify whether LINC01126 and miR-518a-5p were significantly enriched in AGO-containing micro-ribonucleoprotein complexes.
Results: We selected LINC01126, which was the most highly expressed lncRNA, to further verify its functions in periodontitis-induced hypoxia. The expression of LINC01126 was increased in periodontal tissues. In vitro experiment demonstrated that LINC01126 suppressed proliferation, promoted apoptosis and inflammation of hPDLCs under hypoxia via sponging miR-518a-5p. Moreover, we identified HIF-1α acted as a direct target of miR-518a-5p in hPDLCs and LINC01126 promoted periodontitis pathogenesis by regulating the miR-518a-5p/HIF-1α/MAPK pathway.
Conclusion: LINC01126 promotes periodontitis pathogenesis of hPDLCs via miR-518a-5p/HIF-1α/MAPK pathway, providing a possible clue for LINC01126-based periodontal therapeutic approaches.
Keywords: HIF-1α; hypoxia; long non-coding RNA; miR-518a-5p; periodontitis.
© 2020 The Authors. Cell Proliferation published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Long intergenic non-coding RNA 01126 activates IL-6/JAK2/STAT3 pathway to promote periodontitis pathogenesis.Oral Dis. 2025 Jan;31(1):193-205. doi: 10.1111/odi.15033. Epub 2024 Jun 9. Oral Dis. 2025. PMID: 38852165
-
The long non-coding RNA mir155hg promotes NLRP3-inflammasome activation and oxidative stress response in acute lung injury by targeting miR-450b-5p to regulate HIF-1α.Free Radic Biol Med. 2024 Sep;222:638-649. doi: 10.1016/j.freeradbiomed.2024.07.005. Epub 2024 Jul 15. Free Radic Biol Med. 2024. PMID: 39019096
-
MZB1 targeted by miR-185-5p inhibits the migration of human periodontal ligament cells through NF-κB signaling and promotes alveolar bone loss.J Periodontal Res. 2022 Aug;57(4):811-823. doi: 10.1111/jre.13014. Epub 2022 Jun 2. J Periodontal Res. 2022. PMID: 35653494
-
Interactions Between Non-Coding RNAs and HIF-1alpha in the Context of Colorectal Cancer.Biomolecules. 2025 Apr 1;15(4):510. doi: 10.3390/biom15040510. Biomolecules. 2025. PMID: 40305214 Free PMC article. Review.
-
LncRNA in periodontal tissue-derived cells on osteogenic differentiation in the periodontitis field.Oral Dis. 2024 Oct;30(7):4087-4097. doi: 10.1111/odi.14970. Epub 2024 Apr 24. Oral Dis. 2024. PMID: 38655682 Review.
Cited by
-
Androgen-repressed lncRNA LINC01126 drives castration-resistant prostate cancer by regulating the switch between O-GlcNAcylation and phosphorylation of androgen receptor.Clin Transl Med. 2024 Jan;14(1):e1531. doi: 10.1002/ctm2.1531. Clin Transl Med. 2024. PMID: 38214432 Free PMC article.
-
m6A-related long noncoding RNAs predict prognosis and indicate therapeutic response in endometrial carcinoma.J Clin Lab Anal. 2023 Jan;37(1):e24813. doi: 10.1002/jcla.24813. Epub 2022 Dec 16. J Clin Lab Anal. 2023. PMID: 36525280 Free PMC article.
-
Long non-coding RNA ZFY-AS1 represses periodontitis tissue inflammation and oxidative damage via modulating microRNA-129-5p/DEAD-Box helicase 3 X-linked axis.Bioengineered. 2022 May;13(5):12691-12705. doi: 10.1080/21655979.2021.2019876. Bioengineered. 2022. PMID: 35659193 Free PMC article.
-
Analysis of circRNAs profile in TNF-α treated DPSC.BMC Oral Health. 2022 Jul 3;22(1):269. doi: 10.1186/s12903-022-02267-2. BMC Oral Health. 2022. PMID: 35786385 Free PMC article.
-
The Expression and Regulatory Roles of Long Non-Coding RNAs in Periodontal Ligament Cells: A Systematic Review.Biomolecules. 2022 Feb 12;12(2):304. doi: 10.3390/biom12020304. Biomolecules. 2022. PMID: 35204802 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

